Whole Genome Sequencing vs. Targeted Sequencing: A Strategic Guide for Research and Drug Development
This article provides a comprehensive comparison of Whole Genome Sequencing (WGS) and Targeted Sequencing for researchers, scientists, and drug development professionals.
Whole Genome Sequencing vs. Targeted Sequencing: A Strategic Guide for Research and Drug Development
Abstract
This article provides a comprehensive comparison of Whole Genome Sequencing (WGS) and Targeted Sequencing for researchers, scientists, and drug development professionals. It covers foundational principles, genomic region coverage, and variant detection capabilities. The content explores methodological workflows, clinical and research applications in oncology, rare diseases, and infectious diseases, and details cost-benefit analyses and strategies for workflow optimization. A direct comparative analysis evaluates performance, data management, and interpretation challenges, offering evidence-based guidance for selecting the appropriate sequencing approach to maximize efficiency and discovery potential in biomedical research.
Core Principles and Genomic Landscapes: Understanding WGS and Targeted Sequencing
In the field of modern genomics, researchers and clinicians are primarily faced with three powerful sequencing approaches: whole-genome sequencing (WGS), whole-exome sequencing (WES), and targeted sequencing panels. Each method offers a distinct balance of breadth, depth, and cost, making them uniquely suited for different research and clinical applications [1] [2]. The fundamental difference lies in the genomic territory they cover—from the entire 3 billion base pairs of the human genome to a focused selection of genes known to be associated with specific diseases [2].
This guide provides an objective comparison of these technologies, supported by experimental data and detailed methodologies, to inform decision-making for researchers, scientists, and drug development professionals. The choice between these methods is not merely technical but strategic, impacting the depth of analysis, the clarity of results, and the ultimate translational potential of genomic findings in precision medicine.
The following table summarizes the core technical specifications and capabilities of WGS, WES, and targeted panels, providing a foundation for their comparison.
Table 1: Core Technical Specifications of WGS, WES, and Targeted Sequencing
| Feature | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Sequencing Panels |
|---|---|---|---|
| Sequencing Region | Entire genome (coding & non-coding) [2] | Protein-coding exons (~2% of genome) [2] [3] | Selected genes or regions of interest [3] |
| Approximate Region Size | 3 Gb (3 billion base pairs) [2] | > 30 Mb (30 million base pairs) [2] | Tens to thousands of genes [2] |
| Typical Sequencing Depth | > 30X [2] | 50-150X [2] | > 500X [2] |
| Data Output per Sample | > 90 GB [2] | 5-10 GB [2] | Varies with panel size |
| Primary Detectable Variant Types | SNPs, InDels, CNVs, Fusions, Structural Variants (SVs) [2] | SNPs, InDels, CNVs, Fusions [2] [3] | SNPs, InDels, CNVs, Fusions [2] |
| Key Strengths | Comprehensive variant discovery; detection of structural variants and non-coding mutations [3] | Cost-effective focus on known pathogenic variants; good for rare diseases [3] | High depth for sensitive mutation detection; cost-efficient; simplified data analysis [3] |
| Key Limitations | High cost; massive data storage/analysis; interpretation challenges in non-coding regions [3] | Misses non-coding and deep intronic variants; lower sensitivity for structural variants [3] | Limited to known genes; cannot discover novel disease-associated genes [3] |
The hierarchy of genomic coverage is clear: WGS > WES > Targeted Sequencing [2]. WGS provides the most complete picture, while targeted sequencing offers a focused, high-resolution view of pre-defined regions. WES sits in between, capturing a broad swath of the most clinically relevant segments—the exons—where an estimated 85% of known pathogenic variants reside [3].
Experimental Data and Performance Benchmarks
Comparative Analysis in Precision Oncology
A pivotal 2025 study directly compared WES/WGS with transcriptome sequencing (TS) to targeted panel sequencing (TruSight Oncology 500/TruSight Tumor 170) in a clinical setting using samples from 20 patients with rare or advanced tumors [4]. The findings highlight the practical trade-offs between these methods.
Table 2: Comparison of Therapy Recommendations from WES/WGS/TS vs. Panel Sequencing in Oncology
| Metric | WES/WGS with Transcriptome Sequencing (TS) | Targeted Panel Sequencing |
|---|---|---|
| Median Therapy Recommendations per Patient | 3.5 | 2.5 |
| Basis of Recommendations | 176 biomarkers across 14 categories, including complex biomarkers (TMB, MSI, HRD scores), somatic DNA variants, RNA expression, and germline variants. | Limited to the predefined genes and biomarker types covered by the panel. |
| Overlap | Approximately half of the therapy recommendations were identical between both methods. | |
| Unique Value | Approximately one-third of WES/WGS/TS recommendations relied on biomarkers not covered by the panel. | The majority (8 out of 10) of implemented, molecularly-informed therapies were supported by the panel. |
This study demonstrates that while panel sequencing captures most clinically actionable findings, WES/WGS with TS can provide a significant volume of additional therapeutic options, roughly 30-40% more in this cohort, by uncovering complex biomarkers and alterations outside the panel's scope [4].
Comparative Analysis for Mitochondrial DNA
A 2021 study offers a focused comparison specifically for mitochondrial DNA (mtDNA) analysis, sequencing 1499 participants from the Severe Asthma Research Program (SARP) using both WGS and mtDNA-targeted sequencing [5]. The experimental protocol is outlined in the diagram below.
Diagram 1: mtDNA Sequencing Workflow
The study concluded that both methods had a comparable capacity for determining genotypes, calling haplogroups, and identifying homoplasmies (where all mtDNA copies are identical) [5]. However, a key difference emerged in detecting heteroplasmies (a mixture of wild-type and mutant mtDNA within a cell). There was significant variability, especially for low-frequency heteroplasmies, indicating that the sequencing method can influence the detection of these mixed populations [5]. This finding underscores the need for caution when interpreting heteroplasmy data and suggests that targeted sequencing may be sufficient for many mtDNA applications where high-resolution detection of low-level heteroplasmy is not critical.
Essential Research Reagents and Solutions
The execution of genomic sequencing experiments relies on a suite of specialized reagents and tools. The following table details key materials used in the featured experiments.
Table 3: Key Research Reagent Solutions for Sequencing Workflows
| Reagent / Kit / Software | Primary Function | Example Use in Featured Studies |
|---|---|---|
| Kapa Hyper Library Prep Kit | PCR-free library preparation for WGS to reduce amplification bias. | Used in the SARP study for WGS library prep from 500 ng DNA input [5]. |
| REPLI-g Mitochondrial DNA Kit | Whole genome amplification of mtDNA to enrich target regions. | Used for mtDNA-enrichment in the targeted sequencing arm of the SARP study [5]. |
| Nextera XT DNA Library Prep Kit | Rapid library preparation for sequencing from small DNA input. | Used for preparing libraries from mtDNA-enriched samples in the SARP study [5]. |
| BWA (Burrows-Wheeler Aligner) | Aligns sequencing reads to a reference genome. | Used in both the SARP and MASTER studies for aligning reads to the reference genome (rCRS/hg38) [5] [4]. |
| MitoCaller | A likelihood-based method for calling mtDNA variants, accounting for sequencing errors and mtDNA circularity. | The primary variant caller for mtDNA in the SARP study [5]. |
| HaploGrep2 | Tool for determining mtDNA haplogroups from sequencing data. | Used for mtDNA haplogroup classification in the SARP study [5]. |
| Arriba | Software for the rapid discovery of gene fusions from RNA sequencing data. | Used in the reanalysis of the MASTER program data for fusion detection [4]. |
The choice between WGS, WES, and targeted sequencing is not a matter of identifying a single superior technology, but of aligning the tool with the specific research or clinical objective.
For hypothesis-driven research where the genetic targets are well-defined, such as monitoring known cancer drivers, targeted panels offer an efficient, sensitive, and cost-effective solution [3]. For unbiased discovery, the investigation of rare diseases with unknown causes, or the comprehensive assessment of complex biomarkers like TMB and HRD, WGS and WES are indispensable [4] [3]. The continuing decline in sequencing and data storage costs is making WGS increasingly accessible, positioning it as a future first-tier test that can reduce the diagnostic odyssey for many patients [6] [3].
As the field evolves, the integration of artificial intelligence and improved bioinformatics pipelines will be critical for managing and interpreting the vast data generated, particularly by WGS, ultimately unlocking the full potential of precision genomics in research and drug development [7] [3].
Next-generation sequencing (NGS) has revolutionized genomics, but its effectiveness hinges on two critical metrics: sequencing depth and coverage [8] [9]. While often used interchangeably, they represent distinct concepts. Sequencing depth, or read depth, refers to the average number of times a specific nucleotide is read during sequencing (e.g., 30x) [8] [9]. Coverage describes the percentage of the target genome or region that has been sequenced at least once (e.g., 95%) [8] [9].
The choice between Whole Genome Sequencing (WGS) and Targeted Sequencing fundamentally shapes the depth and coverage strategy. WGS aims for comprehensive coverage of the entire genome but typically at a lower, more uniform depth due to cost constraints. Targeted sequencing sacrifices breadth for depth, focusing immense sequencing power on specific regions of interest to detect rare variants with high confidence [1] [10] [8]. This guide objectively compares these approaches, detailing their performance implications through experimental data and standardized methodologies.
Defining the Metrics: A Comparative Framework
The table below summarizes the core differences between Whole Genome and Targeted Sequencing regarding depth, coverage, and their applications.
Table 1: Whole Genome Sequencing vs. Targeted Sequencing - A Comparative Framework
| Aspect | Whole Genome Sequencing (WGS) | Targeted Sequencing |
|---|---|---|
| Scope & Objective | Sequences the entire genome (coding and non-coding regions) to provide an unbiased view and discover novel variants [1] [10]. | Sequences a predefined subset of the genome (e.g., exome, gene panels) to investigate specific, known genetic markers [1] [10]. |
| Typical Depth | 30x - 50x for human genomes [8]. | 50x - 100x for gene mutations; up to 500x-1000x for detecting low-frequency variants in cancer genomics [8]. |
| Coverage Goal | High uniformity across the entire genome, though some complex regions may be challenging to cover [8]. | Very high coverage focused on the targeted regions, ensuring they are comprehensively represented [10] [8]. |
| Primary Applications | Discovery research, novel variant identification, complex disease studies, and de novo genome assembly [10]. | Clinical diagnostics, oncology (e.g., tumor sequencing), and studying inherited disorders with known genetic causes [10] [8]. |
| Cost & Resource Implications | Higher cost due to the extensive sequencing and computational resources required for data analysis [10]. | Generally more cost-effective for focused applications, with simplified data analysis due to reduced data volume [1] [10]. |
Experimental Data and Performance Comparison
Empirical studies directly comparing sequencing platforms highlight the tangible impact of the depth-coverage trade-off on experimental outcomes.
A key study sequenced a mixture of ten HIV clones using both 454/Roche (longer reads) and Illumina (shorter reads) platforms [11]. For a fixed cost, the experimental data demonstrated that short Illumina reads could be generated at much higher coverage, enabling the detection of variants at lower frequencies [11]. However, the assembly of full-length viral haplotypes was only feasible with the longer 454/Roche reads, underscoring the trade-off between high-depth, short-range variant detection and long-range haplotype reconstruction [11].
The quantitative results from such comparative studies can be summarized as follows:
Table 2: Experimental Performance Comparison Based on Platform and Strategy
| Sequencing Strategy | Effective Read Length | Effective Depth/Coverage | Variant Detection Sensitivity | Haplotype Reconstruction Capability |
|---|---|---|---|---|
| Illumina (Short-Read) | Shorter reads (e.g., paired-end 36bp in the cited study) [11]. | Higher coverage for a fixed cost, better for detecting low-frequency single-nucleotide variants (SNVs) [11]. | High sensitivity for detecting low-frequency variants within read length [11]. | Limited to local haplotypes; full-length assembly is generally not feasible [11]. |
| 454/Roche (Long-Read) | Longer reads [11]. | Lower coverage for a fixed cost, but reads connect distant variants [11]. | Lower power for detecting very low-frequency variants due to lower coverage [11]. | High power for assembling global haplotypes and resolving the structure of the virus population [11]. |
Detailed Experimental Protocols
To ensure reproducibility and provide a clear framework for the data discussed, below are detailed methodologies for two common types of experiments cited in comparisons.
Protocol 1: Targeted Sequencing for Variant Detection in Heterogeneous Samples (e.g., Viral Quasispecies or Tumor Biopsies)
This protocol is designed to maximize depth for sensitive variant calling [11] [8].
- Sample Preparation & DNA Extraction: Extract DNA from the sample (e.g., viral RNA converted to cDNA, or genomic DNA from a tumor biopsy). Assess DNA quality and quantity using spectrophotometry or fluorometry.
- Library Preparation - Targeted Enrichment:
- Fragmentation: Fragment the DNA via sonication or enzymatic digestion to a desired size (e.g., 200-500bp) [11].
- Library Construction: Use a kit (e.g., Illumina Genomic DNA sample preparation kit) to repair ends, add 'A' bases, and ligate platform-specific adapters [11].
- Target Enrichment: Employ hybrid capture or PCR amplification to isolate and enrich for the specific genomic regions of interest. This step is crucial for directing sequencing power.
- Sequencing: Load the enriched library onto a high-throughput sequencer (e.g., Illumina). Sequence to a high depth (e.g., ≥500x for low-frequency variants in cancer) using a paired-end protocol to improve mapping accuracy [11] [8].
- Data Analysis:
- Read Mapping: Align the generated reads to a reference genome using a read mapper like Novoalign or SMALT [11].
- Variant Calling: Use specialized software to identify single-nucleotide variants (SNVs) and indels, statistically distinguishing true biological variants from sequencing errors based on the high depth of information [11] [8].
Protocol 2: Whole Genome Sequencing for Comprehensive Variant Discovery
This protocol prioritizes uniform coverage across the entire genome [10] [8].
- Sample Preparation & DNA Extraction: Extract high-quality, high-molecular-weight genomic DNA.
- Library Preparation - Whole Genome:
- Fragmentation: Fragment the DNA randomly into smaller pieces.
- Library Construction: As in Protocol 1, repair ends and ligate adapters without a targeted enrichment step. This creates a library representing the entire genome.
- Sequencing: Sequence the library on an appropriate platform (e.g., Illumina, PacBio) to the desired average depth (e.g., 30x for human WGS). The lack of enrichment leads to a more uniform distribution of reads, albeit at a lower average depth per dollar compared to targeted approaches [8].
- Data Analysis:
- Read Mapping & Assembly: Map all reads to the reference genome. For de novo assembly, use sophisticated bioinformatics tools to reconstruct the genome from the short reads without a reference [10].
- Variant Calling & Annotation: Call variants across the entire genome and annotate their potential functional impact in both coding and non-coding regions [10].
Visualizing the Sequencing Strategy Trade-Offs
The logical relationship between sequencing strategy, its characteristics, and its resulting applications can be visualized in the following workflow.
Diagram: Sequencing Strategy Decision Workflow
The fundamental trade-off between read length and depth of coverage for specific genomic tasks is another critical concept, as demonstrated in the HIV quasispecies study [11].
Diagram: Read Length vs. Depth Trade-Off
The Scientist's Toolkit: Essential Research Reagent Solutions
The following table details key reagents and materials required for the sequencing workflows described in the experimental protocols.
Table 3: Key Reagents and Materials for Sequencing Workflows
| Item | Function | Application Context |
|---|---|---|
| High-Quality DNA Extraction Kit | To isolate intact, pure genomic DNA or cDNA from source material (e.g., blood, tissue, cells). | Fundamental first step for both WGS and Targeted Sequencing [8]. |
| Library Preparation Kit (e.g., Illumina DNA Prep) | Contains enzymes and buffers for DNA end-repair, 'A'-tailing, and adapter ligation to prepare fragments for sequencing. | Core library construction for both WGS and Targeted protocols [11] [8]. |
| Targeted Enrichment Probes/Panels | Biotinylated oligonucleotide probes or primer sets designed to hybridize to and capture specific genomic regions of interest. | Essential for Targeted Sequencing to isolate desired genes/exons before sequencing [10]. |
| Sequence-Specific Adapters & Indexes | Short, known DNA sequences ligated to fragments, allowing for sample multiplexing and binding to the sequencing flow cell. | Required for all NGS protocols on platforms like Illumina [11]. |
| Cluster Generation Reagents | Enzymes and nucleotides used on the sequencer to amplify single DNA molecules into clonal clusters, enabling detection. | Core chemistry for sequencing-by-synthesis platforms like Illumina. |
| Polymerase and Fluorescent Nucleotides | The engine of sequencing; a DNA polymerase incorporates fluorescently-labeled terminator nucleotides during each cycle. | Core chemistry for sequencing-by-synthesis platforms like Illumina. |
The choice between Whole Genome and Targeted Sequencing is a strategic decision governed by the fundamental trade-off between depth and coverage. Whole Genome Sequencing offers an unbiased, comprehensive view of the genome, making it indispensable for discovery research. In contrast, Targeted Sequencing provides a cost-effective, high-depth solution for focused investigations where maximum sensitivity for specific, known variants is required. The experimental data and protocols outlined provide a framework for researchers to make an informed choice, ensuring their sequencing strategy is optimally aligned with their biological questions and clinical objectives.
The choice between whole genome sequencing (WGS) and targeted sequencing (TS) represents a fundamental strategic decision in genetic research and clinical diagnostics. While WGS aims to comprehensively interrogate the entire genome, TS focuses on specific regions of interest with enhanced depth and efficiency [12]. Each approach offers distinct advantages and limitations in detecting various types of genetic variants—including single nucleotide polymorphisms (SNPs), insertions and deletions (indels), copy number variations (CNVs), and structural variations (SVs)—that drive biological processes and disease pathogenesis. This guide provides an objective comparison of the variant detection capabilities of these sequencing methodologies, supported by experimental data and detailed protocols to inform researchers, scientists, and drug development professionals in selecting appropriate strategies for their specific applications.
Comparative Analysis of WGS and Targeted Sequencing
Table 1: Fundamental characteristics of WGS versus targeted sequencing approaches
| Feature | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Panels |
|---|---|---|---|
| Sequencing Region | Entire genome | Protein-coding exons (~1% of genome) | Selected genes/regions of interest |
| Region Size | ~3 Gb | ~30 Mb | Tens to thousands of genes |
| Typical Sequencing Depth | >30X | 50-150X | >500X |
| Approximate Data Output | >90 GB | 5-10 GB | Varies by panel size |
| Detectable Variant Types | SNPs, InDels, CNVs, SVs, fusions | SNPs, InDels, CNVs, fusions | SNPs, InDels, CNVs, fusions |
| Primary Advantage | Comprehensive variant discovery without prior region selection | Balance between coverage and cost for coding regions | Maximum depth for sensitive variant detection in known regions |
Source: Adapted from CD Genomics comparison [2]
Table 2: Performance metrics for variant calling in WGS
| Variant Type | Recall Rate | Precision | Key Limitations |
|---|---|---|---|
| SNVs | >99.9% [13] | >99.9% [13] | Reduced accuracy in repetitive regions [14] |
| Indels (deletions) | Similar to long-read data in nonrepetitive regions [14] | Similar to long-read data in nonrepetitive regions [14] | Significant reduction in recall for insertions >10 bp [14] |
| Indels (insertions >10 bp) | Significantly lower than long-read data [14] | Varies by algorithm [14] | Performance decreases with increasing indel size [14] |
| Structural Variations | Significantly lower in repetitive regions [14] | Similar to long-read in nonrepetitive regions [14] | Particularly challenging for small-intermediate SVs in repetitive elements [14] |
| Copy Number Variants | 97% (NovaSeq X with DRAGEN) [15] | High but platform-dependent [15] | Coverage drops in GC-rich regions affect some platforms [15] |
The fundamental difference between these approaches lies in their scope and depth. WGS provides unbiased coverage across the entire genome, enabling discovery of novel variants in both coding and non-coding regions [2]. In contrast, TS focuses on predetermined genomic regions, achieving much higher sequencing depths that enhance sensitivity for detecting low-frequency variants [12]. This makes TS particularly valuable for applications like tumor sequencing where detection of rare subclones is critical, or for clinical diagnostics where only specific disease-associated genes are of interest [12].
Experimental Protocols for Variant Detection
Whole Genome Sequencing Protocol
Library Preparation and Sequencing The standard WGS protocol begins with quality control of input DNA, typically requiring 100-1000 ng of high-molecular-weight genomic DNA. Library preparation involves fragmentation of DNA to ~350 bp fragments using ultrasonication (e.g., Covaris ultrasonicator) [16]. Following fragmentation, DNA undergoes end repair, A-tailing, and adapter ligation. Libraries are then amplified using cluster generation on a flow cell and sequenced on platforms such as Illumina NovaSeq X Plus using 150 bp paired-end reads, achieving approximately 30-40× coverage [16] [15].
Variant Calling Pipeline Raw sequencing data undergoes base calling to produce raw reads, followed by quality control checks. Quality-filtered reads are aligned to a reference genome (e.g., GRCh38) using BWA-MEM (parameters: mem -t 4 -k 32 -M) [16]. PCR duplicates are marked and removed using SAMTools rmdup [16].
Variant calling employs multiple specialized algorithms:
- SNPs and small InDels: Called using SAMTools mpileup (parameters: -m 2 -F 0.002 -d 1000) with filtering for read depth ≥4 and mapping quality ≥20 [16]
- CNVs: Detected using CNVnator (parameter: -call 100) based on read-depth divergence from reference [16]
- SVs: Identified using BreakDancer for large-scale insertions, deletions, inversions, and translocations based on discordant read pairs and insert size deviations [16]
Functional annotation of variants is performed using tools like ANNOVAR to categorize consequences (exonic, splicing, regulatory etc.) [16].
Targeted Sequencing Protocol
Hybrid Capture-Based Approach The TruSight Rapid Capture kit protocol exemplifies hybrid capture TS. DNA is "tagmented" (fragmented and end-polished using transposons), followed by adapter and barcode ligation [17]. Three to eight libraries are pooled for hybridization with target-specific oligos at 58°C, with two consecutive hybridization cycles to enhance specificity [17]. After capture, libraries are quantified using Bioanalyzer and Qubit assays, diluted to 4 nmol/L, denatured with NaOH, and sequenced with 5% PhiX spike-in for quality control [17].
Amplicon-Based Approach The Ion AmpliSeq protocol represents amplicon-based TS. DNA is amplified in multiple primer pools covering targeted regions, followed by combining PCR products for barcoding and library preparation [17]. Library concentration is measured using TaqMan quantification, adjusted to 40 pmol/L, and loaded onto chips for sequencing [17].
Quality Control and Validation Targeted sequencing requires specific quality metrics:
- On-target rate: Percentage of sequencing data aligning to target regions [2]
- Coverage uniformity: Evenness of coverage across target sites, measured by Fold-80 (additional sequencing needed for 80% of targets to reach mean depth) [2]
- Duplication rate: Percentage of duplicate reads, with lower rates indicating more efficient library complexity [2]
Diagram Title: WGS and TS Experimental Workflows
Performance Assessment and Benchmarking
Reference Materials and Benchmarking Standards
The Genome in a Bottle (GIAB) Consortium developed reference materials for five human genomes, which provide high-confidence "truth sets" of small variants and homozygous reference calls [17]. These materials enable standardized performance assessment across sequencing platforms and analytical pipelines. The GIAB benchmark includes challenging genomic regions such as segmental duplications, low-mappability regions, and repetitive sequences, allowing comprehensive evaluation of variant calling accuracy [15].
Performance metrics follow GA4GH standardized definitions, with sensitivity calculated as TP/(TP+FN) and precision as TP/(TP+FP) [17]. The NIST v4.2.1 benchmark for the HG002 reference genome represents the current gold standard for assessing SNV, indel, and SV calling accuracy [15].
Platform-Specific Performance Characteristics
Table 3: Platform comparison based on benchmarking against GIAB standards
| Platform | SNV Accuracy | Indel Accuracy | Challenging Region Performance |
|---|---|---|---|
| Illumina NovaSeq X | 99.94% vs. NIST v4.2.1 [15] | 22× fewer errors than UG 100 [15] | Maintains high accuracy in GC-rich regions and homopolymers [15] |
| Ultima Genomics UG 100 | 6× more errors than NovaSeq X [15] | Higher error rate, especially in homopolymers >10 bp [15] | Masks 4.2% of genome including challenging regions [15] |
| Long-read Technologies | High accuracy with PacBio HiFi [14] | Superior for insertions >10 bp [14] | Excellent performance in repetitive regions [14] |
Comparative studies reveal that short-read technologies demonstrate excellent SNV and small deletion detection in nonrepetitive regions, with performance comparable to long-read sequencing [14]. However, short-read platforms show significantly lower recall for insertions larger than 10 bp and for SVs in repetitive regions [14]. The performance gap between short and long reads is less pronounced in nonrepetitive regions [14].
Notably, different platforms employ distinct benchmarking strategies. While Illumina typically assesses performance against the complete NIST benchmark including all challenging regions, other platforms may limit evaluation to "high-confidence regions" that exclude problematic genomic areas, potentially inflating apparent accuracy [15].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key reagents and materials for sequencing experiments
| Item | Function | Example Products |
|---|---|---|
| DNA Extraction Kits | Isolation of high-quality genomic DNA | Standard phenol-chloroform, column-based kits |
| Library Prep Kits | Fragmentation, end repair, adapter ligation | TruSight Rapid Capture, Ion AmpliSeq Library Kit |
| Target Enrichment | Capture of specific genomic regions | Inherited Disease Panel Oligos, custom baits |
| Sequencing Kits | Cluster generation and sequencing | NovaSeq X Series 10B Reagent Kit, Ion PGM Hi-Q Chef kit |
| Quality Control Tools | Assessment of DNA and library quality | Bioanalyzer, Qubit assays, TapeStation |
| Reference Materials | Method validation and benchmarking | GIAB DNA aliquots, NIST reference materials |
| Alignment Tools | Mapping reads to reference genome | BWA-MEM, Minimap2 |
| Variant Callers | Detection of genetic variants | GATK, DeepVariant, SAMTools, BreakDancer |
Source: Compiled from multiple experimental protocols [16] [17] [15]
The selection between WGS and targeted sequencing involves strategic trade-offs between comprehensiveness and depth, with significant implications for variant detection capabilities. WGS provides the most complete interrogation of the genome, enabling discovery of novel variants across all genomic regions, but at higher cost and data burden [2]. Targeted sequencing offers cost-effective, deep coverage of specific regions of interest, enhancing sensitivity for low-frequency variants but limiting discovery to predetermined targets [12].
The optimal approach depends on research objectives: WGS excels in discovery-phase studies, identification of non-coding variants, and comprehensive structural variant detection, while targeted sequencing proves superior for clinical applications focusing on known disease genes, detection of low-frequency variants in heterogeneous samples, and resource-constrained settings requiring maximal information from specific genomic regions.
As sequencing technologies continue to evolve, with both short-read and long-read platforms demonstrating rapid improvements, regular benchmarking using standardized reference materials remains essential for accurate performance assessment. Researchers should consider their specific variant detection requirements, particularly regarding variant types and genomic contexts, when selecting between these complementary approaches.
Table of Contents
- Introduction
- A Timeline of Sequencing Costs
- Comparative Sequencing Methodologies
- Methodology: How Sequencing Costs are Calculated
- The Technology Driving Cost Reduction
- The Researcher's Toolkit: Essential Components for Sequencing
- Conclusion & Future Directions
The cost of sequencing a human genome has undergone one of the most dramatic reductions in the history of technology, far outpacing the famed Moore's Law that governed computing progress for decades [18] [19]. This journey from a multi-billion-dollar endeavor to a routine laboratory procedure has fundamentally reshaped biological research and is accelerating the integration of genomics into clinical care. This guide provides an objective comparison of whole-genome sequencing (WGS) against targeted approaches like whole-exome sequencing (WES) and targeted panels, framed within the broader thesis that understanding this cost evolution is critical for selecting the appropriate methodology for research and drug development. The data presented herein consolidates information from leading genomic institutions and recent commercial announcements to offer a clear, data-driven perspective for scientists and researchers.
A Timeline of Sequencing Costs
The following table summarizes the key milestones in the cost of sequencing a human genome, highlighting the accelerated decline with the advent of next-generation sequencing (NGS).
Table 1: Historical and Projected Cost of Sequencing a Human Genome
| Year | Cost (US$) | Notes and Context |
|---|---|---|
| 2001 | ~$100 Million | Cost of the first draft sequence from the Human Genome Project [20]. |
| 2006 | ~$20-$25 Million | Estimated cost using Sanger sequencing technologies prior to NGS [21]. |
| 2008 | ~$1.5 Million | Early NGS begins to significantly outpace Moore's Law [18] [22]. |
| 2015 | ~$4,000 | NHGRI recorded cost for a genome [18] [23]. |
| 2019 | ~$1,000 | NHGRI cost drops below the symbolic $1,000 benchmark [19]. |
| 2022 | ~$500 | NHGRI's final updated benchmark cost [24] [23]. |
| 2023-2024 | ~$100 - $500 | Range of consumable costs claimed for new ultra-high-throughput platforms (e.g., Complete Genomics DNBSEQ-T20x2, Ultima UG100) [19]. |
| 2025 (Projected) | ~$285 | Forecast based on percentage change modeling of NHGRI data [25]. |
It is crucial to distinguish between the often-cited consumable cost (reagents for sequencing) and the total cost of ownership. A 2020 microcosting study in a UK clinical lab found the total cost per rare disease case (a trio) was £7,050, highlighting that consumables were the largest cost component (68-72%), but expenses for equipment, staff, bioinformatics, and data storage are substantial [21]. Furthermore, accessibility and cost vary globally; in Africa, for instance, costs can reach up to $4,500 per genome due to import tariffs and logistical challenges [24].
Comparative Sequencing Methodologies
The choice between WGS, WES, and targeted sequencing involves a fundamental trade-off between the breadth of genomic interrogation, depth of coverage, and cost.
Table 2: Comparison of Whole-Genome, Whole-Exome, and Targeted Sequencing
| Feature | Whole-Genome Sequencing (WGS) | Whole-Exome Sequencing (WES) | Targeted Sequencing Panels |
|---|---|---|---|
| Genomic Target | ~3 billion bases (100% of nuclear DNA) [20] | ~60 million bases (~2% of the genome that are protein-coding exons) [1] | A select number of specific genes or regions known to harbor disease-relevant mutations [1] |
| Sequenceable Variants | SNVs, indels, CNVs, structural variants, regions outside exons [20] [1] | Primarily SNVs and small indels within protein-coding regions [1] | Pre-defined mutations (e.g., "hot-spots") within the panel's scope [1] |
| Sequencing Depth | Typically 30x-50x | Often >100x due to smaller target | Very high depth (often >500x) |
| Key Advantage | Comprehensive, hypothesis-free; captures non-coding variants [1]. | Cost-effective for focused analysis of protein-coding regions; greater depth for lower cost vs. WGS [1]. | Highest depth for sensitive variant detection; lowest cost per sample; often clinically actionable [1]. |
| Key Limitation | Higher cost per sample; massive data storage/analysis; interpretation challenges in non-coding regions [21]. | Misses variants in introns and other non-coding regulatory regions [1]. | Limited to known genes; cannot discover novel disease-associated genes [1]. |
| Relative Cost (Consumables) | $$$ | $$ | $ |
The decision-making workflow for selecting the appropriate sequencing method based on research goals and constraints can be visualized as follows:
Methodology: How Sequencing Costs are Calculated
Accurately determining the cost of sequencing a genome is complex, as different institutions track and account for costs differently [20]. The National Human Genome Research Institute (NHGRI), a primary source for cost benchmarks, makes a critical distinction between 'production' and 'non-production' activities [18].
NHGRI 'Production' Costs (Included in Benchmarks):
- Labor, administration, utilities, reagents, and consumables
- Sequencing instruments and large equipment (amortized over time)
- Informatics directly related to sequence production (e.g., laboratory information management systems, initial base calling)
- Data submission to public databases
- Indirect costs related to the above items [18]
NHGRI 'Non-Production' Costs (Excluded from Benchmarks):
- Downstream bioinformatic analysis (e.g., sequence assembly, variant calling, interpretation)
- Technology development to improve sequencing pipelines
- Quality assessment/control for specific projects
- Data storage for long-term archiving [18]
This distinction explains why the widely cited "$1,000 genome" for consumables was achieved years before the NHGRI's production cost benchmark fell to the same level [19]. For a research budget, the "complete cost" must include the often-overlooked non-production activities, particularly analysis and storage [21].
The Technology Driving Cost Reduction
The precipitous drop in cost since 2007 is directly attributable to the shift from Sanger sequencing to NGS platforms [18] [19]. Sanger methods read DNA sequences in a single, continuous strand, which was slow and expensive for large genomes. NGS technologies, pioneered by companies like Illumina, broke this paradigm by:
- Massive Parallelization: Sequencing millions to billions of DNA fragments simultaneously.
- Short-Read Sequencing: Breaking the genome into small fragments that are sequenced in parallel and computationally reassembled using a reference genome [20].
The current competitive landscape is driving costs down further. As of late 2024, manufacturers are in a "race to the sub-$100 genome," with platforms like the Complete Genomics DNBSEQ-T20x2 and Ultima Genomics UG100 claiming consumable costs of $100 or less per genome, while Illumina's NovaSeq X Plus targets a $200 genome [19]. This competition not only reduces reagent costs but also improves data output and instrument efficiency.
The Researcher's Toolkit: Essential Components for Sequencing
Beyond the sequencing instrument itself, a functional sequencing pipeline requires a suite of reagents, equipment, and computational resources. The following table details the key components.
Table 3: Research Reagent Solutions and Essential Materials for NGS
| Item | Function | Considerations for Implementation |
|---|---|---|
| DNA Extraction Kits | Isolate high-quality, high-molecular-weight DNA from sample sources (e.g., blood, tissue, cells). | Quality and quantity of input DNA are critical for successful library preparation. |
| Library Preparation Kits | Fragment DNA and attach adapter sequences that allow fragments to bind to the sequencing flow cell. | A key cost and time driver. Kits are often platform-specific. Includes reagents for amplification and purification. |
| Sequenceing-by-Synthesis (SBS) Kits | Core consumables containing enzymes, nucleotides, and buffers for the cyclical sequencing reactions on the instrument. | The primary consumable cost. Format (e.g., flow cell, cartridge) is specific to the sequencing platform. |
| Benchtop Sequencer | The instrument that performs the NGS run (e.g., Illumina iSeq 100, NextSeq 2000; Complete Genomics DNBSEQ-G400). | Choice depends on required throughput, data output, and budget [26] [19]. |
| Nucleic Acid Quantitator | Precisely measure DNA concentration (e.g., fluorometric methods) before library prep and sequencing. | Essential for normalizing samples and ensuring optimal loading on the sequencer. |
| Bioinformatics Software | Process raw data (base calling, alignment), identify variants, and perform functional annotation. | Requires significant computational resources and expertise. Licenses can be a recurring cost. |
| Data Storage Solution | Archive massive sequencing files (FASTQ, BAM, VCF). A single WGS can require over 100 GB of storage [22]. | Costs for on-premise servers or cloud storage must be factored into the project budget. |
The landscape of genome sequencing costs has evolved from the astronomical to the accessible, empowering researchers to design studies at a scale once unimaginable. The choice between WGS and targeted approaches is no longer solely dictated by cost but by the specific scientific question, with WGS offering unparalleled comprehensiveness and targeted methods providing deep, cost-efficient interrogation of known regions.
Looking forward, the race to lower costs continues, with the $100 genome now a reality for consumables on the latest platforms [19]. The next frontier will focus on overcoming the remaining challenges: slashing the total cost of ownership by reducing analysis expenses, improving the efficiency of data storage, and developing automated, standardized interpretation pipelines. Furthermore, achieving global equity in genomic innovation will require addressing the high costs and infrastructure barriers in low- and middle-income countries [24]. For the research and drug development community, this ongoing evolution promises to further democratize access to genomic information, accelerating the pace of discovery and the translation of genomics into personalized medicine.
Workflows and Real-World Applications in Research and Clinical Settings
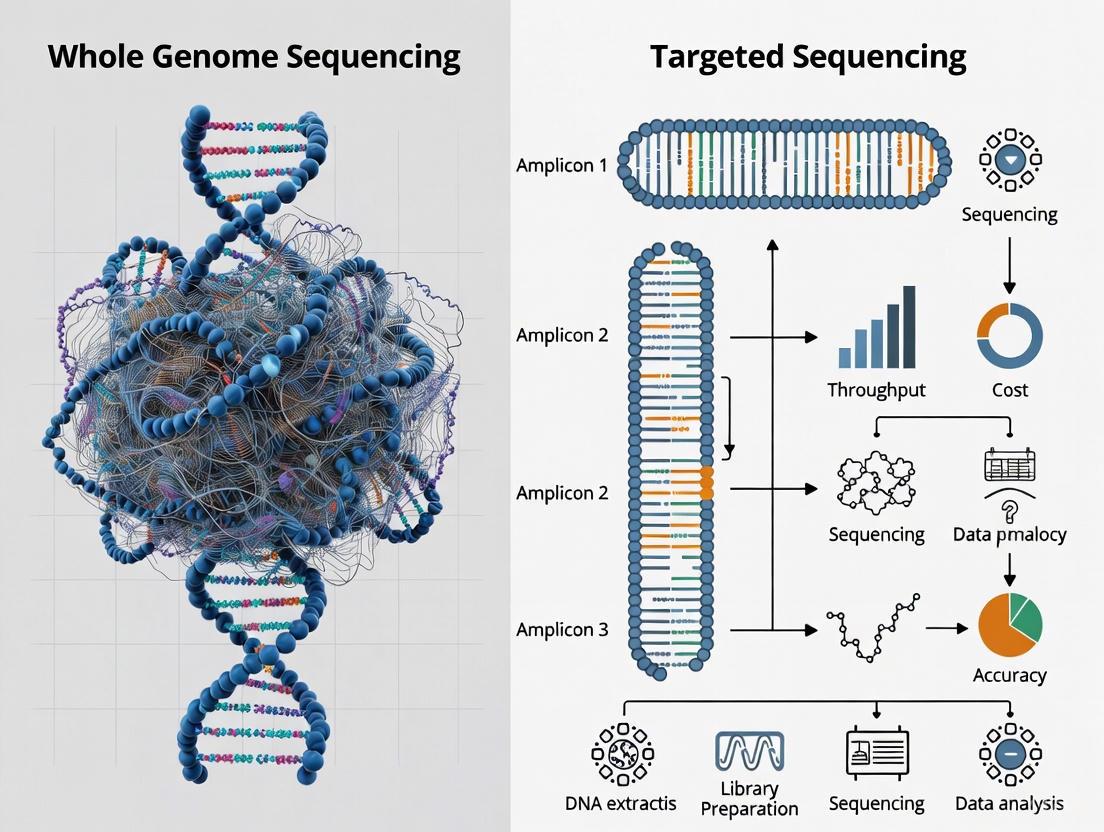
Next-generation sequencing (NGS) has revolutionized genomic research, with whole-genome sequencing (WGS) and targeted sequencing representing two fundamental approaches. WGS aims to sequence the entire genome, approximately 3 billion base pairs in humans, providing an unbiased view of all genetic variants [2]. In contrast, targeted sequencing focuses on specific regions of interest, such as protein-coding exons (whole-exome sequencing, WES) or selected gene panels, enabling deeper coverage of predetermined genomic areas at a lower cost [2] [27]. This guide provides a detailed, step-by-step comparison of these methodologies from initial library preparation through bioinformatics analysis, supported by experimental data to inform researchers, scientists, and drug development professionals.
Methodological Comparison: Library Preparation to Sequencing
Library Preparation Workflows
The initial stages of NGS library preparation share common steps regardless of the eventual sequencing strategy, though with important methodological distinctions.
Core Library Preparation Steps (Common to Both Approaches):
- DNA Fragmentation: Genomic DNA is fragmented to appropriate sizes (typically 300-600 bp) using either mechanical shearing (sonication, nebulization, or focused acoustics) or enzymatic digestion [28]. Mechanical shearing offers more consistent fragment sizes with less bias, while enzymatic digestion requires lower DNA input and enables automation [28].
- End Repair and A-Tailing: The fragmented DNA undergoes end repair to create blunt ends, followed by phosphorylation and 3' adenylation to facilitate adapter ligation [28].
- Adapter Ligation: Platform-specific adapters containing sequencing primer binding sites are ligated to both ends of the DNA fragments [28].
- Library Amplification: PCR amplification is performed to enrich for adapter-ligated fragments, though amplification-free protocols exist to minimize bias [28].
Workflow Divergence for Targeted Sequencing:
After initial library preparation, targeted sequencing requires an additional target enrichment step, which can be accomplished through two primary methods:
- Hybridization Capture: Utilizes biotinylated oligonucleotide probes complementary to target regions. Target-probe hybrids are captured using magnetic beads, while non-target sequences are washed away [28] [27]. This method offers more uniform coverage and is preferred for exome sequencing and detecting rare variants [27].
- Amplicon Sequencing: Employs highly multiplexed PCR with primers designed to amplify specific target regions [28]. This approach requires fewer steps and less input DNA, making it suitable for detecting germline inherited variants and CRISPR editing events [27].
Table 1: Key Differences Between WGS and Targeted Sequencing
| Parameter | Whole Genome Sequencing | Whole Exome Sequencing | Targeted Panels |
|---|---|---|---|
| Sequencing Region | Entire genome (~3 Gb) | Protein-coding exons (~30 Mb) | Selected genes/regions (varies) |
| Region Size | ~3 billion bp | ~30 million bp | Tens to thousands of genes |
| Sequencing Depth | Typically 30X-50X | Typically 50X-150X | Typically >500X |
| Data Output | >90 GB per sample | 5-10 GB per sample | Varies with panel size |
| Detectable Variants | SNPs, InDels, CNV, Fusion, Structural variants | SNPs, InDels, CNV, Fusion | SNPs, InDels, CNV, Fusion |
| Target Enrichment | Not required | Hybridization capture | Hybridization capture or amplicon sequencing |
Sequencing and Data Generation
Following library preparation, samples are loaded onto sequencing platforms. The choice between WGS and targeted sequencing significantly impacts downstream data characteristics:
Coverage and Depth: WGS provides uniform coverage across the entire genome but at relatively lower depth due to cost constraints. Targeted sequencing achieves much higher depth in specific regions, enhancing sensitivity for detecting low-frequency variants [2]. For example, targeted sequencing can detect variants with allele frequencies as low as 1% using hybridization capture without UMIs, and even lower with UMIs [27].
Technical Considerations: The sequencing platform itself introduces technical variability. Studies show that different platforms can yield varying results, with one study reporting only 88.1% concordance for single-nucleotide variants (SNVs) and 26.5% for indels between Illumina and Complete Genomics platforms [29]. Additionally, the amount of input DNA significantly impacts sequencing success, particularly for targeted approaches where insufficient DNA can lead to library preparation failure or adapter contamination [30].
Experimental Data and Performance Comparison
Concordance Studies
Direct comparisons between WGS and targeted sequencing reveal important patterns in variant detection:
Pancreatic Cancer Study: A 2025 paired comparison of WGS and targeted sequencing (Ion Torrent Oncomine Comprehensive Assay Plus) in pancreatic cancer patients demonstrated 81% concordance across all variants and 100% concordance for variants relevant to targeted therapy [31]. Both techniques reliably identified common driver mutations, suggesting that for clinical applications focused on known therapeutic targets, targeted sequencing performs comparably to WGS [31].
Mitochondrial DNA Analysis: A large-scale comparison of WGS and mtDNA-targeted sequencing in 1,499 participants from the Severe Asthma Research Program revealed that both methods had comparable capacity for determining genotypes, calling haplogroups, and identifying homoplasmies [5]. However, significant variability emerged in calling heteroplasmies, particularly for low-frequency variants, highlighting method-specific limitations in detecting mixed populations [5].
Detection Capabilities
The comprehensive nature of WGS enables discovery of variant types typically missed by targeted approaches:
Structural Variants and Non-coding Regions: WGS can identify structural variants (inversions, duplications, translocations) and variations in non-coding regulatory regions that are not covered by targeted panels [2] [28]. These elements may play crucial roles in disease pathogenesis but remain inaccessible to targeted methods.
Rare Variant Detection: While targeted sequencing achieves higher depth for detecting rare variants in specific regions, WGS provides the advantage of genome-wide rare variant discovery without prior knowledge of target regions [27].
Table 2: Performance Comparison Based on Experimental Data
| Performance Metric | Whole Genome Sequencing | Targeted Sequencing |
|---|---|---|
| Variant Concordance | 81-88% with other WGS platforms | 81-100% with WGS for known variants |
| Rare Variant Detection | Genome-wide, but limited by depth | Enhanced in targeted regions (>500X depth) |
| Structural Variant Detection | Comprehensive | Limited to designed targets |
| Heteroplasmy Detection | Variable for low-frequency variants | Variable for low-frequency variants |
| Input DNA Requirements | 500 ng (PCR-free) | 1-250 ng (hybridization capture); 10-100 ng (amplicon) |
Bioinformatics Pipelines and Computational Considerations
Data Processing Workflows
Bioinformatics pipelines for NGS data share fundamental steps but differ in scale and specific approaches:
Primary Data Processing (Common Steps):
- Quality Control: Assessment of raw sequencing data using tools like FastQC to evaluate base quality scores, GC content, and adapter contamination [2].
- Read Alignment: Mapping sequencing reads to a reference genome using aligners such as BWA-MEM or BWA-aln (for reads <70bp) [32]. The choice of aligner affects reproducibility, with some tools like BWA-MEM showing variability when read order is altered [33].
- Duplicate Marking: Identification and flagging of PCR duplicates using tools like Picard MarkDuplicates to prevent variant calling artifacts [32].
- Local Realignment: Correction of misalignments around indels using GATK's IndelRealigner [32].
- Base Quality Score Recalibration: Adjustment of systematic errors in base quality scores using GATK's BaseRecalibrator [32].
Variant Calling and Annotation:
- WGS-Specific Considerations: The comprehensive nature of WGS data requires specialized approaches for detecting structural variants and copy number variations, often employing multiple algorithms [32].
- Targeted Sequencing Considerations: The higher depth in targeted regions enhances sensitivity for somatic variant detection but requires careful handling of off-target reads [2].
- Variant Annotation: Identified variants are annotated with functional information using tools like ANNOVAR to interpret potential biological impacts [2].
Reproducibility and Technical Variability
Bioinformatics tools significantly impact reproducibility, defined as the ability to maintain consistent results across technical replicates [33]. Key considerations include:
Algorithmic Biases: Alignment algorithms may exhibit reference bias, favoring sequences containing reference alleles [33]. Tools employ different strategies for handling multi-mapped reads in repetitive regions, affecting variant calling consistency [33].
Stochastic Variations: Some algorithms incorporate random processes (e.g., Markov Chain Monte Carlo) that can produce different outcomes even with identical input data [33]. Setting random seeds can restore reproducibility in such cases.
Pipeline Selection: No single bioinformatics pipeline has emerged as universally superior. The GDC DNA-Seq pipeline, for instance, implements four separate variant calling pipelines (MuTect2, MuSE, Pindel, VarScan) to provide comprehensive variant detection [32].
Workflow Visualization
Research Reagent Solutions
Table 3: Essential Research Reagents and Tools
| Reagent/Tool | Function | Application Notes |
|---|---|---|
| Hyper Library Preparation Kit (PCR-free) | Library preparation without amplification bias | Ideal for WGS with sufficient DNA input [5] |
| REPLI-g Mitochondrial DNA Kit | Whole mitochondrial genome amplification | Enables mtDNA-targeted sequencing [5] |
| Nextera XT DNA Library Preparation | Transposon-based library preparation | Faster workflow; fragments and tags simultaneously [28] |
| xGen Hybridization Capture Probes | Target enrichment via hybridization | High uniformity; suitable for exome sequencing [27] |
| Oncomine Comprehensive Assay Plus | Targeted cancer panel | Designed for therapeutic biomarker detection [31] |
| BWA Aligner | Sequence alignment to reference genome | Industry standard; BWA-MEM for reads ≥70bp [32] |
| GATK Tools | Variant discovery and genotyping | Provides base quality recalibration, variant calling [32] |
| Picard Tools | SAM/BAM file processing | Handles duplicate marking, file sorting/merging [32] |
The choice between WGS and targeted sequencing involves trade-offs between comprehensiveness and depth. WGS provides unbiased genome-wide coverage, enabling discovery of novel variants and structural variations outside coding regions [2] [28]. Targeted sequencing offers cost-effective, deep coverage of predefined regions, enhancing sensitivity for detecting low-frequency variants and streamlining data analysis [27] [31].
For clinical applications focused on known therapeutic targets, targeted sequencing demonstrates high concordance with WGS while being more resource-efficient [31]. For discovery-oriented research or when investigating non-coding regions, WGS remains the superior approach. Future directions may include hybrid strategies that combine targeted sequencing with low-pass WGS to balance cost and comprehensiveness.
Researchers should select the appropriate method based on their specific research questions, available resources, and desired balance between novel discovery and focused interrogation of genomic regions.
Within the context of a broader thesis comparing whole-genome sequencing to targeted sequencing research, target enrichment stands out as a critical methodological step that enables cost-effective and deep investigation of specific genomic regions. While whole-genome sequencing provides a comprehensive view, its cost and data complexity can be prohibitive for many applications [34]. Targeted sequencing, through enrichment techniques, allows researchers to focus sequencing resources on regions of interest, leading to higher coverage depths, simplified data analysis, and significantly reduced costs [35] [36]. The two most prevalent enrichment methods—hybridization capture and amplicon sequencing—offer distinct advantages and limitations that researchers must carefully consider based on their experimental goals, sample types, and resource constraints. This guide provides an objective comparison of these techniques to inform researchers, scientists, and drug development professionals in selecting the appropriate methodology for their specific applications.
Fundamental Principles and Workflows
Amplicon Sequencing (Multiplex PCR-Based)
Amplicon sequencing utilizes polymerase chain reaction (PCR) to directly amplify specific genomic regions of interest. In this method, multiple primer pairs are designed to flank target sequences and work simultaneously in a multiplexed PCR reaction to create thousands of amplicons [37]. These amplified products are then processed into sequencing libraries by adding platform-specific adapters and sample barcodes [35]. The method is particularly valued for its simplicity and efficiency, enabling rapid library preparation from minimal DNA input—as little as 1 ng in some validated systems [34] [37]. This makes it especially suitable for challenging samples such as formalin-fixed paraffin-embedded (FFPE) tissue, fine needle aspirates, or circulating tumor DNA where sample material is limited [34].
Hybridization Capture-Based Enrichment
Hybridization capture employs biotinylated oligonucleotide probes (baits) that are complementary to targeted genomic regions. The process begins with fragmentation of genomic DNA, followed by adapter ligation and library preparation [35] [37]. The library is then denatured and hybridized with the bait probes in solution. Biotin-labeled probe-target hybrids are captured using streptavidin-coated magnetic beads, while non-target fragments are washed away [37]. The enriched targets are then amplified via PCR before sequencing. This method is particularly advantageous for capturing large genomic regions, with virtually unlimited capacity for targets per panel, making it the preferred approach for whole-exome sequencing and large gene panels [35] [38].
Visual Comparison of Core Workflows
The fundamental differences between these techniques are reflected in their experimental workflows, as illustrated below.
Performance Comparison and Experimental Data
Quantitative Technical Comparison
Extensive evaluations of both methodologies have revealed distinct performance characteristics that directly impact their application suitability. The table below summarizes key comparative metrics derived from published studies.
Table 1: Comprehensive Performance Comparison Between Hybridization Capture and Amplicon Sequencing
| Performance Metric | Hybridization Capture | Amplicon Sequencing | Experimental Context & Notes |
|---|---|---|---|
| On-Target Rate | Variable (typically 50-80%); lower for small panels [39] | Consistently high (>90%); superior for small panels [40] [39] | Amplicon methods achieve higher specificity via primer design [38] |
| Coverage Uniformity | Superior (Fold-80 penalty: ~1.5-2) [40] [41] | Lower uniformity (Fold-80 penalty: >2) [40] | Hybridization demonstrates more even base coverage [40] |
| Sensitivity | <1% variant frequency [35] | <5% variant frequency [35] | Hybridization better for low-frequency variants [35] |
| Sample Input Requirement | 50-500 ng (typical) [35] [37] | 1-100 ng; works with degraded samples [35] [34] [37] | Amplicon superior for limited/scarce samples [37] |
| Variant Detection False Positives/Negatives | Lower noise and fewer false positives [38] | Higher potential for false positives/negatives near primer sites [40] | Amplicon methods can miss variants detected by capture [40] |
| GC Bias | Moderate; better for extreme GC regions [40] | Higher PCR-induced bias [40] [41] | Hybridization handles diverse GC content more effectively [40] |
Practical Implementation Comparison
Beyond technical performance, practical considerations significantly influence method selection for specific research environments and applications.
Table 2: Practical Implementation Characteristics and Application Fit
| Characteristic | Hybridization Capture | Amplicon Sequencing | Implications for Research Use |
|---|---|---|---|
| Workflow Steps | More steps (fragmentation, overnight hybridization, captures) [38] [39] | Fewer steps (multiplex PCR, purification) [35] [38] | Amplicon enables faster turnaround (hours vs. days) [39] |
| Target Capacity | Virtually unlimited (entire exomes) [35] [38] | Flexible, usually <10,000 amplicons per panel [35] [38] | Hybridization preferred for large targets (>1 Mb) [39] |
| Cost Per Sample | Higher (reagents, sequencing) [35] [38] | Generally lower [35] [38] | Amplicon more cost-effective for focused panels [39] |
| Hands-On Time | Significant (multiple handling steps) [39] | Minimal (streamlined workflow) [39] | Amplicon more efficient for high-throughput applications |
| Best-Suited Applications | Whole-exome sequencing, large gene panels, rare variant discovery, cancer research [35] [38] | Genotyping by sequencing, CRISPR validation, germline SNPs/indels, disease-associated variants [35] [38] | Application dictates optimal method selection |
Essential Research Reagent Solutions
Successful implementation of either target enrichment strategy requires specific reagent systems and tools. The following table outlines essential materials and their functions for both methodologies.
Table 3: Essential Research Reagents and Tools for Target Enrichment
| Reagent Category | Specific Examples | Function in Workflow | Method Compatibility |
|---|---|---|---|
| Library Preparation | KAPA HyperPrep, Illumina TruSeq, Ion AmpliSeq | Fragments DNA, adds platform-specific adapters, incorporates sample indices | Both Methods |
| Enrichment Probes/Primers | Agilent SureSelect, IDT xGen, Roche SeqCap, Ion AmpliSeq Primers | Target-specific oligonucleotides for capture or amplification | Method-Specific |
| Capture Materials | Streptavidin-coated magnetic beads | Binds biotinylated probes for target isolation | Hybridization Capture |
| Enzymatic Mixes | Polymerases, ligases, restriction enzymes | Amplifies targets, ligates adapters, digests unused primers | Both Methods (different types) |
| Design Tools | Agilent eArray, Roche HyperDesign, ParagonDesigner | In silico probe/primer design and coverage analysis | Both Methods |
| Quality Control | Agilent Bioanalyzer, Qubit Fluorometer, TapeStation | Assesses DNA quality, quantity, and library fragment size | Both Methods |
Experimental Protocols for Method Evaluation
Standardized Hybridization Capture Protocol
Based on methodologies from comparative studies [42] [40], a representative hybridization capture protocol includes:
DNA Fragmentation: Dilute 1-3 μg genomic DNA and shear to a target peak of 150-300 bp using a focused-ultrasonicator (e.g., Covaris S220) per manufacturer's specifications [40].
Library Preparation: Use a validated library prep kit (e.g., Illumina TruSeq) to repair DNA ends, add platform-specific adapters containing sample barcodes, and perform limited-cycle PCR amplification [40].
Hybridization: Combine the library with biotinylated RNA or DNA probes (e.g., Agilent SureSelect) in hybridization buffer. Incubate at 65°C for 16-24 hours to allow probe-target hybridization [42] [40].
Target Capture: Add streptavidin-coated magnetic beads to bind biotinylated probe-target hybrids. Wash repeatedly with optimized buffers to remove non-specifically bound DNA [37] [40].
Post-Capture Amplification: Elute captured targets from beads and perform 10-14 cycles of PCR to amplify the enriched library for sequencing [40].
Quality Control: Validate library quality using appropriate methods (e.g., Agilent TapeStation) and quantify using fluorometric methods before sequencing [40].
Representative Amplicon Sequencing Protocol
Based on established systems like Ion AmpliSeq [34] [40]:
Panel Design/Primer Pool Preparation: Design primers to flank all targets of interest. For custom panels, use design tools (e.g., Ion AmpliSeq Designer) that leverage algorithms to select optimal primers with minimal interference [34].
Multiplex PCR: Combine 10-250 ng DNA with primer pools (up to 24,000 primer pairs in a single reaction) and robust PCR mix. Amplify with thermal cycling conditions optimized for the specific panel [34] [40].
Primer Digestion: Treat PCR products with enzymes (e.g., FuPa enzyme in Ion AmpliSeq) to partially digest primers and phosphorylate DNA ends in preparation for adapter ligation [34].
Adapter Ligation: Add barcoded adapters to amplicons using ligase enzyme. These adapters contain platform-specific sequences, sample indices, and sequencing primer binding sites [34].
Library Purification: Clean up the final library using magnetic beads to remove enzymes, salts, and unused adapters [34] [39].
Quality Assessment: Evaluate library quality and quantity using appropriate methods (e.g., Agilent High Sensitivity D1K ScreenTapes) before sequencing [40].
Application-Oriented Method Selection Guide
The choice between hybridization capture and amplicon sequencing is primarily driven by research goals, target size, and sample characteristics. The decision pathway below provides a systematic approach to method selection.
Both hybridization capture and amplicon sequencing offer powerful, complementary approaches for target enrichment in next-generation sequencing applications. Hybridization capture excels in applications requiring comprehensive coverage of large genomic regions, superior uniformity, and detection of low-frequency variants. In contrast, amplicon sequencing provides an optimal solution for focused panels, challenging sample types, and high-throughput applications where workflow efficiency, cost-effectiveness, and rapid turnaround are paramount. The choice between these methodologies should be guided by specific research objectives, target characteristics, sample quality, and available resources. As targeted sequencing continues to evolve, both techniques will remain essential tools in the researcher's arsenal, enabling deeper insights into genomic variation and its role in disease and biological processes.
In the evolving landscape of genomic analysis, the choice between whole genome sequencing (WGS) and targeted sequencing is pivotal for research and clinical applications. This guide provides an objective comparison of these technologies, focusing on their performance in gene discovery and variant detection, supported by experimental data and current market trends.
Next-generation sequencing (NGS) has revolutionized genomic research, enabling high-throughput, cost-effective analysis of DNA and RNA [43]. The two primary approaches—whole genome sequencing (WGS) and targeted sequencing—differ fundamentally in scope and application. WGS aims to sequence the entire genome, approximately 3 billion base pairs in humans, providing a comprehensive view of all genetic information, including both coding and non-coding regions [1] [2]. In contrast, targeted sequencing focuses on a curated set of genes or regions of interest, such as the exome (whole-exome sequencing, or WES) or smaller gene panels [2]. While WGS captures the complete genetic blueprint, targeted methods provide deeper coverage of specific regions at a lower cost per sample, making each suitable for distinct research scenarios [1] [2].
Technical Comparison and Performance Data
The technical performance of WGS and targeted sequencing varies significantly across key parameters, influencing their suitability for different research objectives.
Table 1: Key Technical Specifications of Sequencing Approaches
| Parameter | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Panels |
|---|---|---|---|
| Sequencing Region | Entire genome (∼3 billion bases) [2] | Exonic regions only (∼30 million bases) [2] | Selected regions (dozens to thousands of genes) [2] |
| Typical Sequencing Depth | > 30X [2] | 50-150X [2] | > 500X [2] |
| Data Volume per Sample | > 90 GB [2] | 5-10 GB [2] | Varies with panel size [2] |
| Detectable Variant Types | SNPs, InDels, CNVs, Fusions, Structural Variants [2] | SNPs, InDels, CNVs, Fusions [2] | SNPs, InDels, CNVs, Fusions [2] |
| Ability to Discover Novel Genes/Regions | High (comprehensive, hypothesis-free) [43] | Limited to exons [1] | None (restricted to pre-defined panel) [1] |
Table 2: Performance Comparison in Clinical and Research Settings
| Application | WGS Performance & Advantages | Targeted Sequencing Performance & Advantages |
|---|---|---|
| Novel Gene Discovery | Excellent; uncovers variants in non-coding regions and novel structural variants [43] [2]. | Not applicable, as limited to known targets [1]. |
| Rare Variant Detection | Good, but limited by moderate depth. May miss very low-frequency variants [1]. | Excellent; high depth (>500X) enables detection of very low-frequency variants [1] [2]. |
| Clinical Diagnostics (e.g., NICU) | Rapid WGS can provide a hypothesis-free diagnosis in hours [44] [45]. | Targeted panels are efficient when a specific set of disorders is suspected. |
| Non-Invasive Prenatal Testing (NIPT) | Lower failure rates, simpler PCR-free workflow, comprehensive view [46]. | Targeted approaches (e.g., SNP, microarray) analyze limited regions, have more complex workflows [46]. |
| Cost-Effectiveness | Higher per-sample cost; cost-effective for hypothesis-free discovery [47]. | Lower per-sample cost; highly cost-effective for focused, high-volume testing [48]. |
Experimental Data and Protocol Analysis
Case Study: Ultra-Rapid WGS in a Neonatal Intensive Care Unit (NICU)
A groundbreaking study published in 2025 demonstrated the power of WGS in a critical care setting. Researchers from Roche, Broad Clinical Labs, and Boston Children's Hospital set a new world record by sequencing and analyzing a whole human genome in under four hours (3 hours, 57 minutes) [44] [45].
Experimental Protocol:
- Sample Collection: Blood was drawn from NICU infants at the hospital [44].
- Sample Transport: A courier transported the samples to the sequencing facility [44].
- Sequencing Technology: Used Roche's novel Sequencing by Expansion (SBX) technology. This method converts DNA into an expanded surrogate molecule (Xpandomer), generating a high signal-to-noise ratio and enabling extremely fast sequencing [44].
- Data Analysis & Reporting: Sequencing data was continuously analyzed in near real-time. The fastest instance achieved a blood-to-report turnaround time of just 8 hours (6:30 a.m. to 2:30 p.m.) [44].
Results and Implications: The study sequenced 15 genomes, including seven from the NICU. The rapid results, aligning with findings from parallel tests, showcase the potential of WGS to inform urgent clinical decisions, such as avoiding unnecessary procedures and initiating targeted, life-saving treatments for critically ill babies within a single work shift [44] [45].
Protocol for Comparative Technology Assessment
The FDA-led Sequencing Quality Control Phase 2 (SEQC2) project provides a robust framework for comparing sequencing technologies [43].
- Sample Preparation: The study uses well-characterized reference samples, such as the Agilent Universal Human Reference (UHR) from ten cancer cell lines (Sample A) and a cell line from a normal individual (Sample B). Mixtures of A and B in different ratios (e.g., 1:1, 1:4, 4:1) are also created to mimic heterogeneity [43].
- Library Preparation:
- Targeted Sequencing: DNA or RNA libraries are prepared using various targeted panels (e.g., from Agilent, Roche, Illumina) based on hybridization capture principles [43].
- Whole Genome/Transcriptome Sequencing: Libraries are prepared using standard WGS or whole transcriptome (WTS) protocols, including both poly(A) selection and rRNA depletion for RNA [43].
- Sequencing Execution: Libraries are sequenced on multiple short-read (e.g., Illumina) and long-read (e.g., PacBio, Oxford Nanopore) platforms to assess cross-platform performance [43].
- Data Analysis Metrics: The analysis focuses on key performance metrics, including:
- On-target rate: The percentage of sequencing data aligning to the target region.
- Coverage uniformity: The evenness of sequencing depth across target regions.
- Variant calling accuracy: Sensitivity and specificity for calling SNVs, indels, and structural variants.
- Detection of splicing and fusion events: Particularly for RNA sequencing [43].
Market Trends and Adoption Drivers
The market for whole genome and exome sequencing is experiencing exponential growth, projected to grow from $2.02 billion in 2024 to $2.53 billion in 2025, at a compound annual growth rate (CAGR) of 24.8% [6]. This growth is fueled by several key factors:
- Falling Sequencing Costs: Rapid cost compression is making broader panels and even WGS economically feasible in clinical settings. For example, Ultima Genomics reached sub-$100 whole-genome costs in 2024, and Illumina's NovaSeq X lowered per-sample expense by 60% [48].
- Rising Demand in Oncology: Therapy guidelines increasingly require concurrent analysis of multiple genes, prompting labs to replace single-gene tests with large pan-cancer panels. The FDA's classification of NGS tumor-profiling assays as Class II devices in 2024 has further clarified the regulatory path and accelerated adoption [48].
- Expansion in Rare Diseases: While oncology dominates, rare-disease diagnostics is the fastest-growing application segment (24.78% CAGR), driven by newborn genomic-screening pilots and expanded orphan-drug pipelines [48].
- Growth in Non-Invasive Prenatal Testing (NIPT): WGS-based NIPT is gaining traction due to its lower failure rates and simpler, PCR-free workflow compared to targeted approaches like SNP analysis or microarrays [46].
The Scientist's Toolkit: Essential Research Reagents and Materials
The reliability of sequencing experiments depends on the quality of reagents and materials used throughout the workflow.
Table 3: Key Research Reagent Solutions for Sequencing
| Reagent/Material | Critical Function | Application Notes |
|---|---|---|
| Hybridization Capture Probes | Enrich specific genomic regions (e.g., exome or gene panel) from a fragmented DNA library prior to sequencing [2]. | Performance is evaluated by on-target rate, sensitivity, uniformity, and duplication rate [2]. |
| CRISPR-Cas Enrichment | A novel method using guide RNA and Cas enzyme to cleave and enrich specific target regions, offering high specificity and faster design cycles [48]. | Gaining share for its superior performance in GC-rich loci and for structural variant detection with long-read sequencing [48]. |
| Inhibitor-Tolerant Master Mixes | Enzyme mixes resistant to inhibitors found in blood or FFPE (Formalin-Fixed Paraffin-Embedded) samples, enabling direct genotyping without extensive DNA purification [49]. | Crucial for robust clinical sequencing from complex sample types [49]. |
| Library Preparation Kits | Convert extracted DNA or RNA into a format compatible with the sequencing platform through fragmentation, adapter ligation, and amplification [6]. | Kits are often optimized for specific workflows (WGS, WES, or targeted panels) and sample types (e.g., FFPE RNA) [43] [6]. |
| NGS Library Controls | Exogenous spike-in controls (e.g., Virus-Like Particle, VLP) added to the sample to monitor and validate each stage of the assay from extraction to final result [49]. | Essential for comprehensive performance validation and quality assurance in molecular diagnostics [49]. |
The advent of next-generation sequencing (NGS) has revolutionized clinical diagnostics, offering unprecedented capabilities for detecting genetic variations associated with human diseases. Within this landscape, two principal approaches have emerged: whole-genome sequencing (WGS), which aims to determine the order of all nucleotides in an entire genome, and targeted sequencing, which focuses on a select number of specific genes or coding regions known to harbor mutations contributing to disease pathogenesis [1]. While WGS provides a comprehensive view across the entire genome, including non-coding regions, targeted sequencing panels enable deeper sequencing of clinically relevant regions at a lower cost, making them particularly advantageous for clinical applications where specific gene sets are well-characterized [1] [46].
Targeted panels have gained significant traction in clinical settings due to their ability to provide high-depth sequencing for lower cost while delivering greater confidence in low-frequency alterations compared to broader sequencing approaches [1]. These panels typically include clinically actionable genes of interest for diagnostic and theranostic purposes, offering a practical balance between information content, cost-effectiveness, and analytical performance [1]. This guide provides an objective comparison of targeted sequencing panels against alternative genomic approaches, focusing on their performance in oncology, inherited disorders, and infectious disease applications.
Technical Comparison of Sequencing Approaches
Key Methodological Differences
The fundamental distinction between sequencing approaches lies in their scope and enrichment strategies. Whole-genome sequencing employs either de novo assembly, where sequence reads are compared to each other and overlapped to build longer contiguous sequences, or reference-based assembly, which involves mapping each read to a reference genome sequence [50]. In contrast, targeted sequencing panels utilize enrichment techniques such as amplicon-based approaches, which use polymerase chain reaction (PCR) with multiple overlapping amplicons in a single tube to amplify regions of interest, or hybrid capture methods that use oligo probes to capture specific genomic regions [51] [17].
Whole-exome sequencing (WES) represents an intermediate approach, targeting only the exonic regions that compose approximately 2% of the whole genome [1]. Each method offers distinct advantages: WGS provides the most comprehensive collection of an individual's genetic variation; WES enables deeper sequencing of coding regions at lower cost than WGS; and targeted panels achieve the greatest sequencing depth for specific genomic regions, making them ideal for detecting low-frequency variants in clinical settings [1].
Performance Metrics and Experimental Considerations
When evaluating sequencing methodologies, several quality control parameters are essential for assessing data quality. Sequencing depth refers to the ratio of the total number of bases obtained by sequencing to the size of the genome, significantly impacting the completeness and accuracy of variant calling [52]. Coverage represents the proportion of sequenced regions relative to the entire target genome, specifically the ratio of regions detected at least once compared to the total genome [52]. The mapping rate measures the proportion of bases in sequencing data that align to a reference genome, indicating data quality and consistency with the reference [52].
The National Institute of Standards and Technology (NIST) has developed reference materials for five human genomes through the Genome in a Bottle (GIAB) consortium, providing homogeneous DNA aliquots and high-confidence "truth sets" of small variant and homozygous reference calls that enable standardized performance assessment of sequencing methods [17]. These resources allow laboratories to calculate performance metrics using the formula: Sensitivity = TP/(TP+FN), where TP represents true positives and FN represents false negatives [17]. The GIAB materials facilitate understanding of the limitations and optimization of targeted sequencing panels and associated bioinformatics pipelines, with the Global Alliance for Genomics and Health (GA4GH) providing standardized performance metrics and sophisticated variant comparison tools for robust method evaluation [17].
Table 1: Comparative Analysis of Sequencing Methodologies
| Parameter | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Panels |
|---|---|---|---|
| Genomic Coverage | Entire genome (coding + non-coding) | ~2% of genome (exonic regions only) | Select genes/regions of clinical interest |
| Sequencing Depth | Typically lower (30-50x) | Moderate (100-200x) | Very high (500-1000x+) |
| Cost Efficiency | Higher cost | Moderate cost | Lower cost |
| Variant Detection Scope | Comprehensive (SNPs, Indels, CNVs, SVs) | Primarily coding variants | Pre-defined clinically relevant variants |
| Data Volume | Very high (≥100 GB/sample) | Moderate (5-10 GB/sample) | Lower (1-5 GB/sample) |
| Analysis Complexity | High bioinformatics burden | Moderate bioinformatics burden | Streamlined analysis |
| Turnaround Time | Longer | Moderate | Faster |
| Ideal Clinical Use | Rare/undiagnosed diseases, novel gene discovery | Heterogeneous disorders, hypothesis testing | Defined clinical indications, routine testing |
Workflow Comparison
The following diagram illustrates the key procedural differences between whole genome, whole exome, and targeted sequencing approaches:
Figure 1: Sequencing Methodology Workflows. Targeted panels demonstrate streamlined processing with fewer steps compared to broader sequencing approaches.
Targeted Panels in Clinical Oncology
Technology and Performance Metrics
Targeted sequencing panels have transformed molecular oncology by enabling simultaneous assessment of multiple cancer-related genes from various sample types, including formalin-fixed paraffin-embedded (FFPE) tissue and cell-free DNA [51]. These panels utilize multiple overlapping amplicons in a single-tube workflow that can be completed in as little as 2.5 hours to prepare ready-to-sequence libraries, facilitating rapid analysis of tumor samples [51]. The amplicon-based targeted sequencing approach provides confident variant identification at allele frequencies as low as 1%, which is crucial for detecting subclonal populations in heterogeneous tumor samples and identifying driver mutations [51].
The analytical performance of targeted panels is particularly advantageous in oncology applications where detection of low-frequency variants is critical for therapeutic decision-making. For example, the xGen Oncology amplicon panels demonstrate compatibility with Illumina sequencing platforms and offer a fast, easy workflow for both germline and somatic variant identification [51]. These panels employ a PCR1+PCR2 workflow that generates NGS libraries specifically optimized for identifying genetic changes in genes associated with various cancer types, with libraries quantified using conventional methods such as Qubit or Agilent Bioanalyzer and normalized by manual pooling or enzymatic normalization with specialized reagents [51].
Representative Oncology Panels and Their Applications
Commercially available oncology panels target specific genes relevant to particular cancer types or broader pan-cancer applications. The xGen 56G Oncology Amplicon Panel targets 56 genes including ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, and TP53, among others, providing comprehensive coverage of established cancer drivers [51]. The expanded xGen 57G Pan-Cancer Amplicon Panel incorporates an additional gene (TSC2) while maintaining coverage of the core 56-gene set [51]. For more focused applications, disease-specific panels such as the xGen Lung Amplicon Panel (17 genes including EGFR, KRAS, ALK, and MET) and the xGen Colorectal Amplicon Panel (16 genes including APC, KRAS, TP53, and PIK3CA) offer optimized gene selection for particular tumor types [51].
In hematological malignancy profiling, custom NGS panels such as the CleanPlex 25-gene panel for juvenile myelomonocytic leukemia (JMML) demonstrate how targeted sequencing enables differentiated disease classification, risk stratification, and therapeutic decision-making [53]. This amplicon-based targeted sequencing approach provides an ideal balance of cost-effectiveness and analytical performance, allowing researchers to focus specifically on genes related to particular hematologic malignancy subtypes while controlling sequencing costs [53].
Table 2: Representative Targeted Oncology Sequencing Panels
| Panel Name | Number of Genes | Key Genes Covered | Primary Clinical Applications |
|---|---|---|---|
| xGen 56G Oncology | 56 | ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, EGFR, ERBB2, KRAS, TP53 | Broad solid tumor profiling |
| xGen 57G Pan-Cancer | 57 | Includes all 56G genes + TSC2 | Comprehensive pan-cancer analysis |
| xGen Lung Cancer | 17 | EGFR, KRAS, ALK, MET, BRAF, ERBB2, PIK3CA | NSCLC and other lung malignancies |
| xGen Colorectal | 16 | APC, KRAS, TP53, PIK3CA, BRAF, SMAD4 | Colorectal cancer profiling |
| xGen Myeloid | 23 | ASXL1, CALR, CEBPA, DNMT3A, FLT3, IDH1, IDH2, JAK2, NPM1, RUNX1 | Myeloid malignancies (AML, MDS, MPN) |
| xGen BRCA1/BRCA2 PALB2 | 3 | BRCA1, BRCA2, PALB2 | Hereditary breast and ovarian cancer |
| xGen TP53 | 1 | TP53 | Li-Fraumeni syndrome and pan-cancer applications |
| CleanPlex JMML | 25 | Genes frequently mutated in JMML | Juvenile myelomonocytic leukemia |
Experimental Protocol for Targeted Oncology Sequencing
A standardized protocol for targeted sequencing using oncology panels begins with DNA extraction from patient samples, which may include FFPE tissue, frozen specimens, or cell-free DNA from liquid biopsies [51]. For the xGen amplicon panels, the workflow involves: (1) Multiplex PCR where the custom or predesigned panel is combined with the DNA sample to amplify targets of interest; (2) Indexing PCR where samples are amplified with indexing primers to create a functional dual-indexed library; (3) Library normalization using either conventional quantification methods (Qubit, Agilent Bioanalyzer) with manual pooling or enzymatic normalization with xGen Normalase reagents to ensure equal representation of each library in the final sequencing pool [51].
For hybrid capture-based targeted sequencing, such as the TruSight Rapid Capture protocol, the process involves: (1) DNA tagmentation (fragmentation and end-polishing using transposons); (2) Adapter and barcode addition; (3) Library pooling (typically 3-8 libraries); (4) Hybridization with target-specific oligos; (5) Quality assessment using Bioanalyzer high sensitivity DNA chip; (6) DNA quantification with Qubit high sensitivity DNA assay; (7) Library dilution and denaturation; (8) Sequencing with appropriate reagent kits [17]. Throughout this process, incorporation of appropriate controls, including GIAB reference materials, enables performance validation and quality assurance [17].
Targeted Panels for Inherited Disorders
Technology and Applications
Targeted sequencing panels play a crucial role in the diagnosis of inherited disorders by focusing on genes with established associations with monogenic diseases. These panels offer significant advantages over broader sequencing approaches for inherited conditions because they can achieve higher sequencing depths at lower costs while simplifying data interpretation through focused analysis on clinically relevant genes [17]. The higher depth provided by targeted panels is particularly valuable for detecting mosaic variants and for analyzing difficult-to-sequence regions that might be missed by WES or WGS approaches.
The xGen Inherited Disease Research Panel includes targeted assays for conditions such as cystic fibrosis with the xGen CFTR Amplicon Panel, which covers all exons including 5' and 3' UTRs and select intronic regions (1, 12, 22, and 25) of the CFTR gene [51]. Similarly, the xGen Lynch Syndrome Amplicon Panel targets the four mismatch repair genes (MLH1, MSH2, MSH6, PMS2) associated with hereditary non-polyposis colorectal cancer, while the xGen BRCA1/BRCA2 Amplicon Panel and xGen BRCA1/BRCA2 PALB2 Amplicon Panel focus on hereditary breast and ovarian cancer genes [51]. These specialized panels demonstrate how targeted sequencing can be optimized for specific inherited conditions where the genetic etiology is well-established.
Performance Assessment Using Reference Materials
The performance evaluation of targeted panels for inherited disorders benefits from well-characterized reference materials such as those developed by the National Institute of Standards and Technology (NIST) Genome in a Bottle (GIAB) consortium [17]. These reference materials include DNA aliquots from five genomes with high-confidence "truth sets" of small variant and homozygous reference calls that enable standardized assessment of assay performance [17]. The GIAB resources include RM 8398 (GM12878 cell line), RM 8392 (Ashkenazi Jewish trio: GM24143, GM24149, GM24385), and RM 8393 (Chinese ancestry individual: GM24631), providing diverse genomic contexts for test validation [17].
The experimental approach for validating inherited disease panels involves: (1) Sequencing GIAB reference samples using the targeted panel protocol; (2) Variant calling using the laboratory's standard bioinformatics pipeline; (3) Comparison against truth sets using GA4GH benchmarking tools on platforms such as precisionFDA; (4) Calculation of performance metrics including sensitivity, specificity, false positives, and false negatives; (5) Stratified performance analysis by variant type, genome context, and difficult-to-sequence regions [17]. This rigorous validation approach ensures that targeted panels meet the required performance standards for clinical application in inherited disorder diagnosis.
Targeted Approaches in Infectious Disease
Technology and Implementation
Targeted sequencing approaches have found significant application in infectious disease diagnostics, particularly for non-invasive prenatal testing (NIPT) where they compete with whole-genome sequencing methods [46]. The targeted technologies for NIPT include single nucleotide polymorphism (SNP) analysis, microarray analysis, and rolling circle amplification, all of which focus on limited regions of select chromosomes compared to the comprehensive view provided by whole-genome sequencing [46]. Each method employs different biochemical approaches but shares the common principle of selectively analyzing specific genomic regions rather than the entire genome.
In SNP-based NIPT, cell-free DNA is amplified by PCR using specific SNP targets, followed by sequencing and analysis of allele distributions to determine parent-child genetic differences and infer copy number variations [46]. Microarray-based approaches involve amplification of cell-free DNA fragments by PCR, fluorescent probing, and hybridization to complementary sequences on microarrays, with deviations in expected fluorescent counts indicating aneuploidy [46]. Rolling circle amplification targets specific cell-free DNA fragments that bind to circular templates and replicate by a rolling mechanism, with replication products fluorescently labeled and counted to detect deviations indicating aneuploidy [46]. These targeted methods generally involve more complex workflows with additional steps and increased amplification compared to whole-genome sequencing approaches [46].
Performance Comparison in Infectious Disease Applications
In NIPT applications, whole-genome sequencing technology demonstrates performance advantages over targeted methods, including consistently lower failure rates and higher informativeness of results [46]. The PCR-free sample preparation used with whole-genome-sequencing-based NIPT simplifies laboratory workflow, reduces assay complexity, and significantly improves turn-around time compared to targeted approaches [46]. Furthermore, whole-genome sequencing NIPT technology offers superior scalability to accommodate growing laboratory needs [46].
For other infectious disease applications, targeted panels provide focused analysis of pathogen-specific genes or resistance markers. While the search results do not provide extensive details on infectious disease panels, the principles of targeted sequencing similarly apply—focusing on known virulence factors, resistance genes, or species-specific markers to enable efficient pathogen identification and characterization. The sample processing and library preparation workflows for infectious disease targeted panels generally follow similar principles to oncology and inherited disorder applications, with optimization for the specific challenges of microbial detection and quantification in clinical specimens.
Successful implementation of targeted sequencing in clinical research requires specific reagents, reference materials, and computational tools. The following table summarizes key resources mentioned in the search results that facilitate robust targeted sequencing applications:
Table 3: Essential Research Reagents and Resources for Targeted Sequencing
| Resource Category | Specific Examples | Function and Application |
|---|---|---|
| Reference Materials | NIST GIAB RM 8398, RM 8392, RM 8393 [17] | Standardized DNA aliquots with truth sets for assay validation and performance metrics |
| Targeted Panels | xGen Oncology Amplicon Panels [51] | Predesigned gene sets for cancer research with optimized coverage |
| Targeted Panels | CleanPlex Custom NGS Panels [53] | Customizable targeted sequencing assays with ultra-high multiplex PCR |
| Library Prep Kits | TruSight Rapid Capture kit [17] | Hybrid capture-based target enrichment for inherited disease sequencing |
| Library Prep Kits | Ion AmpliSeq Library Kit 2.0 [17] | Amplicon-based target enrichment for inherited disease analysis |
| Normalization Reagents | xGen Normalase reagents [51] | Enzymatic normalization for balanced library representation |
| Quality Control Tools | Agilent Bioanalyzer [51] [17] | Microfluidic analysis of library fragment size distribution and quality |
| Quantification Methods | Qubit fluorometric quantification [51] [17] | Accurate DNA and library concentration measurement |
| Bioinformatics Tools | GA4GH Benchmarking tools [17] | Standardized variant comparison and performance metric calculation |
| Analysis Platforms | precisionFDA [17] | Cloud-based platform for method validation and comparison |
Decision Framework for Sequencing Methodology Selection
The choice between whole genome sequencing, whole exome sequencing, and targeted panels depends on multiple factors including clinical context, research objectives, and practical considerations. The following decision pathway provides a structured approach to methodology selection:
Figure 2: Sequencing Methodology Decision Pathway. This framework guides selection based on clinical needs and practical constraints.
Targeted sequencing panels represent a powerful approach for clinical molecular diagnostics when the genetic basis of disease is well-characterized and defined gene sets provide clinically actionable information. Their advantages include higher sequencing depth for detecting low-frequency variants, lower cost compared to comprehensive sequencing approaches, faster turnaround times, and simplified data analysis and interpretation [51] [1]. However, these advantages come at the expense of comprehensive genomic coverage, potentially missing novel genetic associations or variants in genes not included on the panel [1].
Whole-genome sequencing remains the most comprehensive approach for novel gene discovery and detection of variants in non-coding regions, while whole-exome sequencing provides a balanced solution for conditions with significant genetic heterogeneity where targeted panels may be too restrictive [1] [50]. The future of clinical sequencing will likely involve continued refinement of targeted panels for specific clinical indications, combined with appropriate use of broader sequencing approaches when clinical presentation suggests genetic etologies beyond currently characterized gene-disease associations. As sequencing technologies evolve and costs decrease, the relative advantages of each approach will continue to shift, requiring ongoing evaluation of the optimal strategy for specific clinical and research applications.
The integration of next-generation sequencing (NGS) into pharmaceutical research has fundamentally transformed the drug development pipeline, enabling a shift from traditional one-size-fits-all approaches to precision medicine. By decoding the genetic underpinnings of disease and individual variations in drug response, sequencing technologies provide critical insights from initial target identification through clinical trials and into post-market pharmacovigilance [54] [55]. The choice of sequencing strategy—comprehensive whole-genome sequencing (WGS) or focused targeted sequencing—represents a fundamental strategic decision with significant implications for cost, data complexity, and clinical applicability.
Each approach offers distinct advantages: WGS provides an unbiased view of the entire genome, while targeted sequencing delivers deep, cost-effective coverage of clinically actionable regions [1]. This guide objectively compares these methodologies within the context of drug development, providing researchers and scientists with performance data, experimental protocols, and practical frameworks for selecting the optimal approach to advance therapeutic discovery and personalized medicine.
Technical Comparison: Whole Genome vs. Targeted Sequencing
Fundamental Methodological Differences
Whole-genome sequencing aims to determine the order of all nucleotides (A, C, G, T) across an entire genome, capturing both coding and non-coding regions. This comprehensive view enables identification of genetic variants—including single nucleotide variants (SNVs), insertions, deletions, and copy number variations (CNVs)—anywhere in the genome, including introns and regulatory regions that can influence gene expression and disease [1]. The typical workflow involves fragmenting the entire genome, sequencing all fragments, and computationally reassembling these into a complete genomic sequence.
In contrast, targeted sequencing panels focus on a predetermined set of genes or genomic regions known to harbor mutations contributing to disease pathogenesis or drug metabolism. These panels typically include clinically actionable genes related to specific therapeutic areas, such as oncology, cardiology, or pharmacogenomics [1]. By concentrating sequencing power on specific regions of interest, targeted approaches achieve significantly higher depth of coverage (often 500x-1000x compared to 30x-60x for WGS), enhancing sensitivity for detecting low-frequency variants present in heterogeneous samples like tumors [55].
Performance and Application Comparison
Table 1: Technical and Performance Characteristics of Sequencing Approaches
| Parameter | Whole Genome Sequencing | Targeted Sequencing Panels |
|---|---|---|
| Genomic Coverage | Entire genome (coding + non-coding) | Select genes/regions (typically 1-5 Mb) |
| Sequencing Depth | 30x-60x (standard clinical) | 500x-1000x (common for tumors) |
| Primary Applications in Drug Development | Novel target discovery, biomarker identification, comprehensive genomic profiling | Clinical trial patient stratification, pharmacogenomic testing, routine clinical genotyping |
| Variant Detection Capability | SNVs, indels, CNVs, structural variants, intronic variants | High-sensitivity detection of known SNVs, indels in targeted regions |
| Data Volume per Sample | ~100 GB (raw data) | ~1-5 GB (varies with panel size) |
| Turnaround Time (incl. analysis) | Several days to weeks | 1-3 days for results |
| Cost per Sample (approx.) | Higher ($1000-$5000 clinical grade) | Lower ($200-$1000 depending on panel) |
The selection between these approaches involves clear trade-offs. While WGS provides unprecedented comprehensiveness, this comes with substantial data management challenges, higher costs, and more complex interpretation requirements, particularly for variants of unknown significance in non-coding regions [1] [55]. Targeted sequencing offers practical advantages in clinical settings where specific, known variants guide therapeutic decisions, such as in oncology where panels focus on genes with established roles in cancer pathogenesis and treatment response [55].
Sequencing in Action: Drug Development Workflow
From Genome to Medicine: A Sequencing-Enabled Pipeline
The following workflow illustrates how different sequencing approaches integrate into key stages of modern drug development, from initial discovery through clinical application.
Diagram 1: Sequencing approaches mapped to the drug development pipeline. WGS dominates early discovery phases, while targeted sequencing is preferred for clinical application.
Application-Specific Methodologies
Target Identification and Biomarker Discovery (WGS-focused)
Experimental Protocol: Novel Cancer Gene Discovery
- Sample Collection: Obtain tumor and matched normal tissue from cohorts of patients with specific cancer types (e.g., 100-500 samples).
- Library Preparation: Use PCR-free library preparation methods to reduce bias, particularly in GC-rich regions [46].
- Sequencing: Perform whole-genome sequencing at minimum 30x coverage for normal samples and 60x for tumor samples to adequately detect somatic variants.
- Bioinformatic Analysis:
- Alignment to reference genome (GRCh38) using BWA-MEM or similar tools
- Somatic variant calling with multiple callers (GATK, Mutect2, VarScan)
- Structural variant detection (Manta, Delly)
- Copy number alteration analysis (ASCAT, Sequenza)
- Validation: Confirm findings using orthogonal methods (Sanger sequencing, digital PCR) in independent cohorts.
This approach has identified novel therapeutic targets across cancer types, including previously unrecognized driver mutations in non-coding regions [55].
Clinical Trial Patient Stratification (Targeted Sequencing-focused)
Experimental Protocol: Oncology Trial Enrichment
- Panel Design: Select 50-500 gene regions known to be altered in the cancer type of interest, including genes associated with drug response (e.g., EGFR, ALK, BRCA1/2, KRAS).
- Library Preparation: Use hybrid capture-based target enrichment systems (e.g., Illumina Nextera, Agilent SureSelect) with dual-indexed adapters to enable sample multiplexing.
- Sequencing: Sequence on benchtop platforms (Illumina MiSeq, Ion GeneStudio S5) to high depth (500x minimum) to detect low-frequency clones.
- Variant Calling: Use targeted bioinformatics pipelines with amplicon-aware alignment and duplicate marking.
- Interpretation: Classify variants according to established guidelines (e.g., AMP/ASCO/CAP tiers) to determine clinical actionability.
Targeted approaches enable efficient patient selection for clinical trials based on molecular profiles, as demonstrated in trials matching PARP inhibitors to BRCA-mutated cancers [55].
Essential Research Reagent Solutions
The successful implementation of sequencing in drug development requires carefully selected reagents and platforms optimized for specific applications.
Table 2: Essential Research Reagents and Platforms for Sequencing Applications
| Reagent Category | Specific Examples | Function in Workflow | Application Considerations |
|---|---|---|---|
| Library Prep Kits | Illumina Nextera Flex, Agilent SureSelect, Ion AmpliSeq | Fragment DNA and add platform-specific adapters | PCR-free kits reduce bias for WGS; amplicon-based enable high-multiplexing for targeted |
| Target Enrichment | IDT xGen Pan-Cancer Panel, Thermo Fisher Oncomine | Capture specific genomic regions of interest | Hybrid capture vs. amplicon-based; panel content should reflect therapeutic area |
| Sequencing Platforms | Illumina NovaSeq X, Thermo Fisher Ion GeneStudio S5, PacBio Revio | Generate raw sequencing data | Throughput, read length, cost per sample dictate platform choice |
| Enzymes & Buffers | High-fidelity polymerases, fragmentation enzymes | Amplify and process nucleic acids | Enzyme fidelity critical for variant detection; stability important for reproducibility |
| Bioinformatics Tools | GATK, Sentieon, Fabric Genomics | Variant calling, annotation, interpretation | Automated clinical interpretation platforms accelerate reporting |
The selection of appropriate reagents directly impacts data quality, with targeted panels requiring careful design to ensure coverage of clinically relevant regions while WGS demands high-quality input DNA and minimal amplification bias [56] [55].
Pharmacogenomics: Bridging Genetics to Drug Response
Genetic Determinants of Drug Metabolism and Efficacy
Pharmacogenomics (PGx) represents one of the most mature clinical applications of sequencing in drug development, focusing on how inherited genetic variations influence individual responses to medications. Key genetic polymorphisms in drug metabolism enzymes and transporters (ADME genes) contribute substantially to pharmacokinetic and pharmacodynamic variability [54]. Well-characterized examples include:
- CYP2C19 variants associated with bleeding risk during clopidogrel therapy
- DPYD variants correlated with severe toxicity from 5-fluorouracil or capecitabine
- TPMT polymorphisms linked to thiopurine-induced myelosuppression
- UGT1A1*28 allele associated with irinotecan-induced gastrointestinal toxicity
- SLCO1B1*5 variant increasing risk for simvastatin toxicity [54]
These established gene-drug relationships form the foundation for clinical pharmacogenomic testing and are increasingly integrated into drug labels and treatment guidelines issued by regulatory agencies including the FDA and EMA [54].
Analytical Approaches for Pharmacogenomic Discovery
Experimental Protocol: DMET Array and NGS Integration
- Genotyping Platform: Utilize the DMET (Drug Metabolism Enzymes and Transporters) Plus microarray platform or targeted NGS panels covering 1,936 FDA-recognized markers relevant to drug metabolism.
- Sample Processing: Extract DNA from blood or saliva samples, quantify, and process according to platform specifications.
- Data Generation: Hybridize samples to arrays or sequence using targeted approaches with appropriate controls.
- Bioinformatic Analysis:
- Implement quality control filters for call rates and sample contamination
- Annotate variants using PharmGKB and CPIC databases
- Perform association analyses between genetic variants and drug response phenotypes
- Apply machine learning algorithms to identify polygenic determinants of drug response
- Clinical Interpretation: Classify variants according to functional impact (e.g., poor, intermediate, extensive, or ultrarapid metabolizer phenotypes) [54].
This integrated approach has expanded our understanding of complex polygenic influences on drug response beyond single gene-drug interactions, enabling more comprehensive prediction of drug efficacy and toxicity risk [54].
Market Landscape and Future Directions
The sequencing market continues to evolve rapidly, with the global NGS market projected to grow from $10.27 billion in 2024 to $73.47 billion by 2034, representing a compound annual growth rate of 21.74% [57]. This expansion is particularly pronounced in the genomic biomarkers segment, expected to reach $17 billion by 2033, largely driven by oncology applications that currently account for 35.1% of the genomic biomarkers market [58].
Several converging trends are shaping the future of sequencing in drug development:
- Multiomic Integration: Combining genomic data with transcriptomic, epigenetic, and proteomic datasets to build more comprehensive models of disease biology and therapeutic response [56].
- AI-Enhanced Analytics: Machine learning and artificial intelligence are being deployed to identify complex patterns in large genomic datasets, accelerating biomarker discovery and variant interpretation [56].
- Decentralization of Sequencing: Technological advances are making sequencing more accessible beyond central laboratories, enabling point-of-care genomic testing in diverse clinical settings [56].
- Direct-to-Consumer Expansion: Growing public accessibility to genomic testing is increasing patient engagement with genetic information and creating new opportunities for recruitment into clinical trials [47].
These developments are collectively advancing the field toward more personalized, predictive, and preemptive therapeutic strategies across a widening spectrum of diseases.
The choice between whole-genome and targeted sequencing approaches represents a strategic decision with significant implications for drug development programs. Whole-genome sequencing offers unparalleled comprehensiveness for novel target discovery and comprehensive biomarker identification, particularly valuable in early research phases exploring uncharted biological territory. In contrast, targeted sequencing provides cost-effective, deep coverage of established genomic regions, making it ideal for clinical trial enrichment, pharmacogenomic testing, and routine molecular profiling in validated therapeutic contexts.
As sequencing technologies continue to evolve—with costs declining, platforms improving, and analytical methods becoming more sophisticated—the integration of genomic information throughout the drug development pipeline will increasingly become standard practice. The most successful drug development programs will strategically leverage both approaches at appropriate stages, using WGS for exploratory discovery and targeted methods for clinical development and application, ultimately accelerating the delivery of more effective, safer, and personalized therapeutics to patients.
Strategic Selection and Cost-Efficiency Optimization
For researchers embarking on a genomics project, one of the most critical decisions is whether to cast a wide net across the entire genome or to focus deeply on specific regions of interest. This guide provides an objective comparison between whole-genome sequencing (WGS) and targeted sequencing, offering a data-driven framework to help you select the optimal approach for your research goals.
Next-generation sequencing (NGS) offers multiple paths for genetic analysis, each with distinct advantages and trade-offs. The choice between them hinges on the specific research question, budget, and desired data output.
Whole-Genome Sequencing (WGS) determines the order of all the nucleotides (A, C, G, T) in an organism's entire genome. This allows for the detection of genetic aberrations—including single nucleotide variants (SNVs), insertions, deletions, and copy number variants (CNVs)—anywhere in the genome, including the non-coding introns [1].
Whole-Exome Sequencing (WES) is a focused approach that sequences only the exomes, the 1-2% of the genome composed of exons that code for proteins [1].
Targeted Sequencing uses panels to sequence a select number of specific genes or coding regions known to harbor mutations relevant to a particular disease, such as cancer or inherited disorders [1] [59]. This method achieves the highest sequencing depth (number of times a given nucleotide is sequenced) for a lower cost, which is critical for identifying low-frequency variants [1].
The table below summarizes the core characteristics of each method.
Table: Core Characteristics of Major Sequencing Approaches
| Feature | Whole-Genome Sequencing (WGS) | Whole-Exome Sequencing (WES) | Targeted Sequencing Panels |
|---|---|---|---|
| Target Region | Entire genome (~3 billion bases) | All protein-coding exons (~1-2% of genome) | Selected genes/regions of interest |
| Coverage Depth | Lower (typically 30x-50x) | Higher than WGS | Highest (500x-1000x or more) [59] |
| Variant Detection | Comprehensive; SNVs, indels, CNVs, SVs, in coding and non-coding regions | Primarily coding SNVs and indels | Focused on known or suspected mutations in the panel |
| Key Advantage | Unbiased discovery of novel variants | Cost-effective focus on functional exons | Maximum depth for detecting rare variants; simplest data analysis [1] [59] |
| Primary Limitation | Higher cost per sample; complex data management and analysis | Misses non-coding and structural variants | Limited to pre-defined content; cannot discover novel variants outside the panel [1] |
Performance and Cost Comparison
Selecting a sequencing strategy involves balancing cost, data quality, and the ability to answer the research question. The following data provides a quantitative basis for this decision.
Sequencing Accuracy and Coverage
A critical performance metric is variant-calling accuracy. One internal evaluation compared two modern WGS platforms—the Illumina NovaSeq X Series and the Ultima Genomics UG 100—using the National Institute of Standards and Technology (NIST) v4.2.1 benchmark [15]. The study highlighted that the NovaSeq X Series demonstrated superior accuracy when assessed against the entire genome benchmark, while the UG 100 platform's accuracy was measured against a "high-confidence region" (HCR) that excludes 4.2% of the genome where its performance is less reliable [15].
Table: Variant Calling Performance Against Full NIST v4.2.1 Benchmark
| Performance Metric | Illumina NovaSeq X Series | Ultima Genomics UG 100 Platform |
|---|---|---|
| SNV Errors | 1x (Baseline) | 6x more errors [15] |
| Indel Errors | 1x (Baseline) | 22x more errors [15] |
| Excluded Genome Regions | 0% | 4.2% (UG "High-Confidence Region") [15] |
| Excluded ClinVar Variants | 0% | 1.0% [15] |
| Performance in Homopolymers | Maintains high indel accuracy | Indel accuracy decreases significantly in homopolymers >10 bp [15] |
The regions excluded by targeted analyses can be biologically significant. The UG HCR, for instance, excludes pathogenic variants in 793 genes and misses 1.2% of pathogenic variants in the well-known BRCA1 tumor suppressor gene [15]. Similarly, targeted approaches may struggle with GC-rich sequences, leading to loss of coverage in disease-related genes like B3GALT6 (linked to Ehlers-Danlos syndrome) and FMR1 (causes fragile X syndrome) [15].
Cost and Operational Considerations
The cost of sequencing a whole human genome has plummeted from approximately $100 million in 2001 to just over $500 in 2023 in the United States [24]. However, actual costs can vary significantly based on location, import tariffs, reagent availability, and logistics. In Africa, for example, costs can reach up to $4,500 per genome [24].
For targeted sequencing, the overall cost is lower, but the key economic principle is that cost per sample decreases significantly as sample throughput increases [60]. Pilot data from the Genomics Costing Tool (GCT) illustrates this relationship across different scenarios and platforms.
Table: Cost per Sample Across Different Operational Scenarios (USD)
| Sequencing Platform | Validation Scenario | Optimization Scenario | Scale-up Scenario |
|---|---|---|---|
| Illumina | $241 | $216 | $162 |
| Oxford Nanopore (ONT) | $252 | $227 | $159 |
| Parameters | Annual throughput: 600 samples | Different instrument, same throughput | Same instrument, higher throughput |
Data adapted from GCT pilot exercises [60]
Experimental Protocols and Methodologies
The reliability of sequencing data is fundamentally linked to the laboratory and computational methods used. Below is a detailed protocol from a published study that directly compared WGS and targeted sequencing.
Protocol: Paired Comparison of WGS and Targeted-seq for mtDNA
A 2021 study compared WGS and mtDNA-targeted sequencing (targeted-seq) using 1,499 samples from the Severe Asthma Research Program (SARP) to analyze mitochondrial DNA [5].
Sample Preparation:
- DNA Source: Whole blood samples from all participants [5].
- WGS Library Prep: 500 ng of DNA was used with the Kappa Hyper Library Preparation Kit (PCR-free). Sequencing was performed on the Illumina HiSeq X with 150 bp paired-end reads [5].
- Targeted-seq Library Prep: 20 ng of DNA was digested with enzymes to reduce nuclear DNA. The whole mitochondrial genome was amplified using the REPLI-g mitochondrial DNA kit (QIAGEN). The library was prepared with the Nextera XT DNA Library Prep Kit (Illumina) and sequenced on an Illumina MiSeq System with 151 bp paired-end reads [5].
Bioinformatic Analysis:
- Read Alignment: Raw sequencing data from both methods were aligned to the revised Cambridge Reference Sequence (rCRS) of the mitochondrial genome using BWA (v0.7.12) [5].
- Variant Calling: Mitochondrial DNA variants (heteroplasmies and homoplasmies) were called using MitoCaller, a likelihood-based method that accounts for sequencing error rates and the circularity of the mtDNA genome [5].
- Variant Classification: A site was called as:
- Homoplasmy: if the alternative allele frequency (AAF) was >95%.
- Heteroplasmy: if the AAF was between 5% and 95%.
- Reference: if the AAF was <5% [5].
Key Finding: The study concluded that targeted-seq and WGS have a comparable capacity to determine genotypes and call haplogroups and homoplasmies. However, there was significant variability in calling heteroplasmies, particularly for low-frequency variants, indicating that researchers should be cautious when comparing heteroplasmies from different sequencing methods [5].
Protocol: Adaptive Sampling for Target Enrichment
A novel method called adaptive sampling, available on Oxford Nanopore Technologies (ONT) sequencers, redefines targeted sequencing by performing enrichment or depletion during the sequencing run, with no need for special library preparation [61].
Workflow:
- Library Preparation: Standard, PCR-free library prep (e.g., using the Ligation Sequencing Kit) is performed on the entire DNA sample, preserving long fragments and native DNA modifications [61].
- Run Setup: In the MinKNOW software, the researcher provides a BED file containing the genomic coordinates of targets to be enriched or depleted [61].
- Real-Time Selection: As each DNA strand enters a nanopore, its initial sequence is basecalled and compared against the target list.
- If it matches a target of interest (or is not a region for depletion), sequencing continues.
- If it is not a target, the software reverses the pore voltage to eject the molecule, allowing a new one to be sequenced [61].
Advantages: This method avoids PCR bias, provides long-read data, and allows for dynamic, software-based updates to target regions without changing wet-lab protocols [61]. Our analysis finds this method is particularly useful for enriching large, complex panels, entire chromosomes, or depleting abundant DNA (e.g., host DNA in microbial samples) [61].
A Framework for Selecting Your Approach
Use the following decision tree to identify the most appropriate sequencing method for your project based on its primary goal. This framework synthesizes the performance and cost data to guide your strategy.
Research Reagent Solutions
The following table details key reagents and kits used in the featured experiments, providing a starting point for your own project planning.
Table: Essential Research Reagents for Sequencing Workflows
| Reagent / Kit Name | Function / Application | Compatible Platform(s) |
|---|---|---|
| Kapa Hyper Library Preparation Kit | PCR-free library preparation for WGS to minimize bias [5]. | Illumina |
| REPLI-g Mitochondrial DNA Kit | Whole mitochondrial genome amplification for targeted mtDNA sequencing [5]. | Any (Pre-sequencing) |
| Nextera XT DNA Library Preparation Kit | Rapid library prep for small genomes (e.g., mtDNA) and amplicon sequencing [5]. | Illumina |
| Illumina DNA Prep with Enrichment | A targeted sequencing solution for genomic DNA from tissue, blood, saliva, and FFPE samples [59]. | Illumina |
| Ligation Sequencing Kit | Standard PCR-free library prep for Oxford Nanopore sequencing, preserving long reads and base modifications [61]. | Oxford Nanopore |
| DesignStudio / AmpliSeq for Illumina | Online tools for designing custom targeted enrichment or amplicon sequencing panels [59]. | Illumina |
Maximizing Cost-Efficiency Through Sample Throughput and Platform Selection
Next-generation sequencing (NGS) has revolutionized genomic research, yet selecting the optimal approach requires careful consideration of cost, throughput, and analytical objectives. The fundamental choice between whole genome sequencing (WGS), whole exome sequencing (WES), and targeted sequencing represents a critical trade-off between the comprehensiveness of data and resource efficiency. For researchers and drug development professionals, maximizing cost-efficiency involves matching the sequencing strategy to specific research questions while leveraging technological advances that have dramatically reduced sequencing costs from billions of dollars per genome to under $1,000 in just two decades [20].
This guide provides an objective comparison of sequencing approaches, focusing on how platform selection and experimental design impact cost-efficiency for various research scenarios. We present structured experimental data and methodological details to inform decision-making for genomics research programs.
Technical Comparison of Sequencing Approaches
Core Methodologies and Genomic Coverage
The three primary sequencing approaches differ fundamentally in genomic regions targeted, data output, and applications:
Whole Genome Sequencing (WGS) sequences the entire genome, encompassing both coding (exonic) and non-coding regions. The human genome comprises approximately 3 billion base pairs (3 GB) [2]. WGS provides the most comprehensive variant detection capability, including single nucleotide variants (SNVs), insertions/deletions (Indels), copy number variations (CNVs), and structural variations (SVs) [1].
Whole Exome Sequencing (WES) specifically targets protein-coding regions (exons), which constitute approximately 1% of the human genome (about 30 million base pairs) [2]. The exome includes approximately 180,000 exons that are captured through hybridization methods prior to sequencing [2].
Targeted Sequencing Panels focus on selected genes or genomic regions of known or suspected functional significance, typically ranging from a few dozen to a thousand genes [2]. These panels operate on either hybridization capture or multiplex amplicon sequencing principles and provide the most focused approach [2].
Table 1: Comparison of Key Technical Parameters Across Sequencing Approaches
| Parameter | Whole Genome Sequencing | Whole Exome Sequencing | Targeted Panels |
|---|---|---|---|
| Sequencing Region | Entire genome (~3 Gb) [2] | All exons (>30 Mb) [2] | Selected regions (tens to thousands of genes) [2] |
| Typical Sequencing Depth | >30X [2] | 50-150X [2] | >500X [2] |
| Data Volume per Sample | >90 GB [2] | 5-10 GB [2] | Varies by panel size |
| Detectable Variant Types | SNPs, InDels, CNV, Fusion, SV [2] | SNPs, InDels, CNV, Fusion [2] | SNPs, InDels, CNV, Fusion [2] |
| Key Applications | Comprehensive variant discovery, structural variant analysis, novel biomarker identification [1] | Coding variant identification, Mendelian disorder research, cancer genomics [2] | High-sensitivity mutation detection in known genes, clinical diagnostics, therapeutic targeting [1] |
Cost and Throughput Considerations
Sequencing costs vary significantly based on the approach, with targeted methods offering substantial savings for focused research questions. While WGS provides the most comprehensive data, it generates approximately 9-18 times more data than WES (90 GB vs. 5-10 GB per sample) [2], impacting both sequencing costs and downstream data storage and computational requirements.
The relationship between sequencing depth and cost is a critical factor in experimental design. Targeted sequencing achieves much higher depth (>500X) for the same cost compared to WES (50-150X) or WGS (>30X) [2], enabling more confident detection of low-frequency variants. This makes targeted approaches particularly cost-effective for applications requiring high sensitivity, such as detecting somatic mutations in cancer or heteroplasmy in mitochondrial DNA [5].
Recent platform developments continue to drive down costs while increasing throughput. The 2025 sequencing landscape includes Illumina's NovaSeq X Series, which promises to generate more than 20,000 whole genomes per year, and emerging technologies like Roche's Sequencing by Expansion (SBX) scheduled to launch in 2026 [62]. These advances make higher-throughput WGS more accessible, potentially changing the cost-benefit calculations for large-scale studies.
Experimental Data and Performance Comparison
Direct Method Comparison in Mitochondrial DNA Analysis
A 2021 study directly compared WGS and targeted sequencing for mitochondrial DNA (mtDNA) analysis using 1,499 participants from the Severe Asthma Research Program (SARP) [5]. This paired comparison provides valuable insights into the practical performance differences between these approaches.
Table 2: Performance Comparison of WGS vs. Targeted Sequencing for mtDNA Analysis
| Performance Metric | Whole Genome Sequencing | Targeted Sequencing | Implications |
|---|---|---|---|
| Genotype Determination | High accuracy | Comparable to WGS | Both methods reliable for basic variant calling |
| Haplogroup Calling | Effective | Comparable capacity | Either method suitable for phylogenetic studies |
| Homoplasmy Detection | Effective | Comparable capacity | Consistent performance for high-frequency variants |
| Heteroplasmy Detection | Variable, especially for low-frequency variants [5] | Large variability for low-frequency variants [5] | Caution required for low-frequency heteroplasmies |
| Sample Input Requirements | 500 ng DNA for PCR-free library prep [5] | 20 ng DNA after mtDNA enrichment [5] | Targeted approach more suitable for limited samples |
| Library Preparation Method | Kapa Hyper Library Preparation Kit (PCR-free) [5] | Nuclear DNA digestion + whole mtDNA amplification [5] | Targeted method requires specialized enrichment |
The study revealed that while both methods had comparable capacity for determining genotypes and calling haplogroups and homoplasmies, there was "large variability in calling heteroplasmies, especially for low-frequency heteroplasmies" [5]. This finding highlights the importance of matching the sequencing method to the specific variant types of interest, particularly for detecting low-frequency variants where both methods showed limitations.
Platform Performance Comparison: DNBSEQ vs. Illumina
A comprehensive 2025 study evaluated structural variation (SV) detection performance across sequencing platforms, analyzing eight DNBSEQ and two Illumina whole-genome sequencing datasets of the NA12878 reference sample [63]. The research applied 40 different SV detection tools to assess comparative performance across five SV types: deletions (DELs), duplications (DUPs), insertions (INSs), inversions (INVs), and translocations (TRAs).
Table 3: SV Detection Performance Comparison Between DNBSEQ and Illumina Platforms
| SV Type | Average Count (DNBSEQ) | Average Count (Illumina) | Size Correlation | Sensitivity Correlation | Precision Correlation |
|---|---|---|---|---|---|
| DELs | 2,838 [63] | 2,676 [63] | 0.97 [63] | 0.83 [63] | 0.91 [63] |
| DUPs | 1,490 [63] | 1,664 [63] | 0.85 [63] | 0.91 [63] | 0.80 [63] |
| INSs | 1,117 [63] | 737 [63] | 0.92 [63] | 0.96 [63] | 0.89 [63] |
| INVs | 422 [63] | 239 [63] | 0.88 [63] | 0.85 [63] | 0.84 [63] |
| TRAs | 2,793 [63] | 2,878 [63] | Not assessed | Not assessed | Not assessed |
The study concluded that "the performance of SVs detection using the same tool on DNBSEQ and Illumina datasets was highly consistent," with correlations greater than 0.80 for key metrics including number, size, precision, and sensitivity [63]. This demonstrates that for SV detection, both platforms offer comparable performance, enabling researchers to base platform selection on factors such as cost, throughput, and availability.
Experimental Protocols and Methodologies
Workflow for Whole Exome Sequencing
The standard WES workflow comprises three main stages: library preparation, sequencing, and bioinformatics analysis [2]. Each stage contains critical steps that impact both cost and data quality:
Library Preparation Stage:
- Sample Processing and DNA Extraction: Isolating high-quality DNA from biological samples
- Quantification: Precisely measuring DNA concentration to ensure adequate input material
- Library Construction: Fragmenting DNA and adding platform-specific adapters
- Hybridization Capture: Using probe-based hybridization to enrich exonic regions
- Amplification: PCR amplification of captured libraries
- Quality Control: Assessing library quality and quantity before sequencing [2]
Sequencing Stage:
- Utilization of either short-read (Illumina, DNBSEQ) or long-read (Oxford Nanopore, PacBio) platforms
- Adjustment of sequencing depth based on research requirements (typically 50-150X for WES) [2]
Bioinformatics Analysis:
- Quality Control: FastQC for assessing sequencing data quality
- Alignment: BWA for mapping reads to the reference genome
- Variant Calling: GATK for identifying genetic variations
- Annotation: ANNOVAR for adding functional information to variants [2]
Targeted Sequencing Protocol for Influenza A Virus
An optimized 2025 workflow for influenza A virus (IAV) surveillance demonstrates how targeted approaches can maximize cost-efficiency for specific applications [64]. The protocol utilizes a multisegment RT-PCR (mRT-PCR) approach with modified conditions to enhance amplification of all eight IAV segments:
Key Methodological Improvements:
- Use of LunaScript RT Master Mix Kit with modified primer ratios (1:4 ratio of MBTuni-12 to MBTuni-12.4 primers at final molarity of 0.5 μM)
- Optimized reverse transcription conditions: 2 minutes at 25°C followed by 30 minutes at 55°C
- Implementation of Q5 Hot Start High-Fidelity DNA Polymerase for improved PCR fidelity
- Introduction of dual-barcoding approach for Oxford Nanopore platform enabling multiplexing of at least eight samples per library barcode [64]
This optimized protocol demonstrated improved recovery of all eight genomic segments, particularly the larger polymerase genes (PB1, PB2, PA) that are challenging to amplify from low viral load samples [64]. The method maintained robustness across avian, swine, and human IAV samples, illustrating how protocol optimization can enhance throughput and cost-efficiency for targeted sequencing applications.
Probe Evaluation Criteria for Targeted Sequencing
For hybridization-based targeted approaches (including WES), careful probe evaluation is essential for cost-efficient experimental design:
Key Evaluation Metrics:
- On-Target Rate: Percentage of sequencing data aligning to the target region; higher rates indicate less wasted sequencing [2]
- Coverage: Percentage of target regions sequenced at a given depth; typically reported as "10X coverage of 90%" [2]
- Homogeneity: Evenness of coverage across target regions; measured by Fold-80 (additional sequencing needed for 80% of targets to reach average depth) [2]
- Duplication Rate: Percentage of duplicate reads; lower rates indicate more efficient capture [2]
These metrics directly impact cost-efficiency, as higher on-target rates, more uniform coverage, and lower duplication rates reduce the sequencing depth required to confidently call variants, thereby lowering per-sample costs.
Visualization of Sequencing Selection Workflows
Diagram 1: Decision workflow for selecting cost-efficient sequencing strategies
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagent Solutions for Sequencing Workflows
| Reagent/Category | Specific Examples | Function in Workflow | Application Notes |
|---|---|---|---|
| Library Preparation Kits | Kapa Hyper Library Preparation Kit [5] | PCR-free library construction for WGS | Minimizes amplification bias in whole genome studies |
| Target Enrichment Systems | REPLI-g mitochondrial DNA Kit [5] | Whole mitochondrial genome amplification | Enables targeted mtDNA sequencing from limited input |
| Reverse Transcription Kits | LunaScript RT Master Mix Kit [64] | cDNA synthesis for RNA virus sequencing | Optimized for multisegment amplification in viral surveillance |
| High-Fidelity Polymerases | Q5 Hot Start High-Fidelity DNA Polymerase [64] | Accurate amplification in targeted protocols | Critical for maintaining sequence fidelity in amplification |
| Target Capture Probes | Various commercial exome panels [2] | Hybridization-based enrichment of target regions | Key determinant of on-target rate and coverage uniformity |
| Sequencing Platforms | Illumina NovaSeq X, DNBSEQ-T1+, Oxford Nanopore [62] | Massive parallel sequencing | Platform choice affects read length, accuracy, and throughput |
| Nucleic Acid Extraction Kits | NucleoMag VET kit, QIAamp Viral RNA Mini Kit [64] | Nucleic acid isolation from various sample types | Critical first step affecting downstream data quality |
Maximizing cost-efficiency in sequencing requires careful matching of methodological approaches to research objectives. Targeted sequencing provides the highest sensitivity for known genomic regions at the lowest cost, making it ideal for clinical diagnostics and focused research questions. Whole exome sequencing offers a balanced approach for coding variant discovery, while whole genome sequencing delivers comprehensive variant detection at higher cost but provides the most complete genomic inventory.
Platform selection continues to evolve, with DNBSEQ platforms demonstrating comparable performance to Illumina for variant detection [63], potentially increasing competition and cost-efficiency. Emerging technologies like Roche's SBX and Illumina's 5-base chemistry promise further enhancements in throughput and informational content [62].
Experimental design considerations—including appropriate sequencing depth, sample multiplexing strategies, and careful probe selection—remain critical factors in optimizing cost-efficiency. By aligning technical capabilities with research goals and leveraging the latest platform advancements, researchers can maximize sample throughput and data quality within budget constraints, accelerating discoveries in genomics and personalized medicine.
In the evolving landscape of next-generation sequencing (NGS), the choice between whole-genome sequencing (WGS) and targeted sequencing approaches represents a fundamental strategic decision for researchers. While WGS provides a comprehensive, base-by-base view of the entire genome, targeted sequencing enables researchers to focus on specific genomic regions of interest with significantly greater depth and cost-efficiency [1] [65]. The performance of targeted sequencing hinges critically on the effectiveness of the capture probes used to enrich genomic material, with three metrics serving as paramount indicators of probe quality: on-target rate, uniformity, and specificity [41] [2]. This guide provides an objective comparison of probe performance evaluation, presenting experimental data and methodologies essential for researchers, scientists, and drug development professionals to make informed decisions in their genomic studies.
Sequencing Approaches: A Comparative Framework
Targeted sequencing has emerged as an important routine technique in both clinical and research settings, offering advantages including high confidence and accuracy, reasonable turnaround time, relatively low cost, and reduced data burdens compared to whole-genome approaches [12]. The three primary NGS approaches—whole-genome sequencing (WGS), whole-exome sequencing (WES), and targeted panels—occupy distinct positions in the research and clinical workflow, each with characteristic strengths and limitations [1] [2].
Table 1: Comparison of Primary DNA Sequencing Approaches
| Parameter | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Sequencing Panels |
|---|---|---|---|
| Sequencing Region | Entire genome (~3 Gb in humans) | Protein-coding exons (~30-60 Mb) | Selected genes/regions (customizable size) |
| Region Size | ~3 billion base pairs | ~30 million base pairs | Tens to thousands of genes |
| Typical Sequencing Depth | 30X-60X | 50X-150X | >500X (often 1000X+) |
| Data Volume | >90 GB per sample | 5-10 GB per sample | Minimal (depends on panel size) |
| Detectable Variants | SNVs, InDels, CNVs, SVs, regulatory elements | SNVs, InDels, CNVs | SNVs, InDels, CNVs, fusions (panel-dependent) |
| Primary Applications | Discovery research, novel variant identification, de novo assembly | Disease-specific research, clinical sequencing | Clinical diagnostics, liquid biopsy, inherited disease, oncology |
| Cost Considerations | Highest ($$$) | Medium ($$) | Lowest ($) |
Targeted sequencing panels specifically focus on a selected number of genes or genomic regions known to be associated with disease pathogenesis, enabling deeper sequencing at lower costs while providing greater confidence for clinical applications [1] [12]. For profiling challenging clinical samples with lower tumor content or degraded DNA quality—such as circulating tumor DNA (ctDNA) and formalin-fixed paraffin-embedded (FFPE) samples—targeted sequencing provides substantially greater sequencing depth (1000× or higher) compared to non-NGS techniques [12]. This enhanced depth is critical for detecting rare variants present in a small fraction of cells and can detect variant allele frequencies (VAF) as low as 0.1–0.2% in minimal residual disease monitoring [12].
Core Metrics for Probe Performance Evaluation
The effectiveness of targeted sequencing approaches depends fundamentally on the performance of enrichment probes. The following three metrics serve as the primary indicators of probe quality and efficiency.
On-Target Rate
The on-target rate measures the specificity of the target enrichment experiment and is defined as the percentage of sequencing data that aligns with the intended target region [41] [2]. This metric can be calculated in two ways: percent bases on-target (the number of bases mapping to the target region) and percent reads on-target (the percentage of sequencing reads overlapping the target region) [41]. A higher on-target rate indicates strong probe specificity, high-quality probes, and efficient hybridization-based target enrichment [41]. Off-target data represents wasted sequencing resources and cannot be utilized in subsequent analyses, making this metric particularly important for cost-efficient study design [2].
Low on-target rates typically result from suboptimal probe design, poorly optimized protocols, problems during library preparation or hybrid capture, or low-quality reagents [41]. To improve on-target rates, researchers should invest in well-designed, high-quality probes, robust reagents, and validated, reliable enrichment methods [41].
Uniformity of Coverage
Uniformity of coverage describes how evenly sequencing reads are distributed across targeted regions in the genome [41] [66]. Ideally, all targeted regions should receive similar sequencing depth, but in practice, some regions capture more efficiently than others due to variations in GC content, probe binding efficiency, and other factors [41]. This metric is critically important for variant detection, as regions with insufficient coverage may miss true variants [66].
The Fold-80 base penalty metric quantifies coverage uniformity by describing how much additional sequencing is required to bring 80% of the target bases to the mean coverage level [41]. A perfect uniformity score would be 1.0, indicating that 80% of bases already reach mean coverage without additional sequencing [41]. Values higher than 1 indicate uneven coverage, with greater values representing poorer uniformity. For example, a Fold-80 value of 2 indicates that twice as much sequencing is needed for 80% of reads to reach the mean coverage [41]. The Fold-80 base penalty provides information about the capture efficiency of probes in a panel, which is impacted by both probe design and probe quality [41].
Specificity
Specificity refers to a probe's precision in capturing intended genomic regions without off-target effects [2]. High-specificity probes minimize cross-hybridization with non-target regions that share sequence homology with targets [2]. This metric is particularly important when targeting genes with pseudogenes or highly homologous family members, where non-specific enrichment can compromise data quality and variant calling accuracy [67].
In practical terms, specificity directly influences the efficiency of a sequencing experiment—probes with higher specificity generate more usable data per sequencing dollar, as less capacity is wasted on off-target regions [2]. Techniques to enhance specificity include careful probe design that avoids repetitive regions, optimization of hybridization conditions, and the use of blocker oligonucleotides to prevent non-specific binding [68].
Experimental Assessment of Probe Performance
Comparative Experimental Protocol
A comprehensive study comparing commercially available target enrichment methods provides valuable insights into experimental protocols for evaluating probe performance [68]. Researchers from the DNA Sequencing Research Group (DSRG) designed an experiment where identical genomic samples and target regions were provided to leading probe manufacturers for independent analysis using their respective platforms [68].
Table 2: Experimental Parameters for Probe Performance Comparison
| Parameter | Agilent SureSelect | Roche NimbleGen SeqCap EZ |
|---|---|---|
| Enrichment Method | Solution-based hybridization | Both array-based and solution-based hybridization |
| Probe Type | RNA probes | DNA probes |
| Target Region Size | ~3.5 Mb total | ~3.5 Mb total |
| Sample Type | Human genomic DNA (Coriell Institute) | Human genomic DNA (Coriell Institute) |
| Replication | Duplicate experiments | Duplicate experiments |
| Sequencing Platform | Illumina Genome Analyzer IIx | Illumina Genome Analyzer IIx |
| Analysis Parameters | Design coverage, sensitivity, specificity, uniformity, reproducibility | Design coverage, sensitivity, specificity, uniformity, reproducibility |
The target region totaled 3.5 Mb and included 31 individual genes with varying chromosome locations, locus sizes (1,565–423,700 bp), GC content, and alternative transcript numbers, plus a contiguous 2-Mb region of chromosome 11 [68]. This design enabled researchers to evaluate probe performance across genomically diverse regions.
Key Findings from Comparative Studies
Analysis of the resulting sequencing data revealed several important trends in probe performance. In the targeted regions, researchers detected 2546 SNPs with the NimbleGen samples compared to 2071 with Agilent's technology [68]. When analysis was limited to regions that both companies included as baits, the number of SNPs was approximately 1000 for each, with each platform identifying a small number of unique SNPs not detected by the other [68].
Overall, coverage variability was higher for the Agilent samples across the targeted regions [68]. The success of enrichment was found to be highly dependent on the design of the capture probes, with both platforms demonstrating strengths in different genomic contexts [68].
Advanced Probe Technologies and Methodologies
Linked Target Capture (LTC)
Innovations in probe technology continue to emerge, addressing limitations of traditional approaches. Linked Target Capture (LTC) represents a novel targeted sequencing library preparation method that replaces typical multi-day target capture workflows with a single-day, combined "target-capture-PCR" workflow [69]. This approach uses physically linked capture probes and PCR primers and is expected to work with panel sizes from 100 bp to >10 Mbp [69].
The LTC method uses Probe-Dependent Primers (PDPs) consisting of non-extendable DNA capture probes linked 5' to 5' with a low melting-temperature universal primer complementary to a portion of the ligated adapter [69]. When bound to their targets, the probes bring the universal primer into close proximity with the universal priming site on the template, increasing the reaction rate of primer binding and initiating polymerase extension [69]. This method demonstrates high on-target read fractions due to repeated sequence selection in the target-capture-PCR step, thereby lowering sequencing costs [69].
Hybridization Capture vs. Amplicon-Based Approaches
Targeted NGS libraries can be enriched using two primary techniques: hybridization capture or amplicon-based enrichment [67]. Each approach offers distinct advantages for specific applications:
Hybridization Capture uses molecules complementary to target regions as probes to select target molecules from the sample [67]. These capture probes can be immobilized on solid substrates (array-based format) or used directly in solution [67]. Solution-based hybridization—the more common contemporary approach—uses biotinylated probes to hybridize with targets, which are then isolated and purified using streptavidin magnetic beads [67].
Amplicon-Based Enrichment employs carefully designed highly-multiplexed PCR to amplify regions of interest from DNA or cDNA samples [67]. This approach offers several distinct advantages: it requires lower sample input (enabling work with limited sources like FFPE tissue or circulating tumor DNA), can better discriminate between highly homologous genomic regions through precise primer design, and more effectively detects known insertions and fusion events that might disrupt hybridization capture [67].
Diagram 1: Workflow for Probe Performance Evaluation. This diagram illustrates the comprehensive process for assessing key probe metrics across different enrichment technologies.
Essential Research Reagents and Solutions
Successful probe evaluation and targeted sequencing require specific laboratory reagents and computational tools. The following table outlines essential resources for researchers designing probe performance studies.
Table 3: Essential Research Reagent Solutions for Probe Evaluation
| Category | Specific Products/Tools | Function/Application |
|---|---|---|
| Commercial Probe Systems | Agilent SureSelect, Roche NimbleGen SeqCap, IDT xGen | Customizable target enrichment systems with established performance characteristics |
| Library Prep Kits | KAPA Target Enrichment, Illumina DNA Prep | Robust library preparation workflows that minimize GC-bias and optimize yield |
| Sequencing Platforms | Illumina NovaSeq X Series, Ultima UG 100 | High-throughput sequencing with varying performance characteristics across genomic regions |
| Analysis Tools | Picard CollectHsMetrics, SAMtools, FastQC, BWA, GATK | Calculation of key metrics including on-target rate, Fold-80 penalty, and coverage uniformity |
| Reference Materials | Genome in a Bottle (GIAB) Consortium, Coriell Institute samples | Characterized reference materials for assay development, quality control, and validation |
| Quality Metrics | Depth of coverage, GC-bias, duplicate rate, fold-80 base penalty | Comprehensive assessment of sequencing performance and probe efficiency |
Platform-Specific Performance Considerations
Recent comparative analyses of sequencing platforms reveal important implications for probe performance evaluation. The Illumina NovaSeq X Series demonstrates higher variant calling accuracy compared to the Ultima Genomics UG 100 platform, with 6× fewer single nucleotide variant (SNV) errors and 22× fewer indel errors when assessed against the full NIST v4.2.1 benchmark [15]. Notably, the UG 100 platform employs a "high-confidence region" (HCR) that excludes 4.2% of the genome from analysis, including challenging regions such as homopolymers, repetitive sequences, and areas with low coverage [15]. This masking approach potentially impacts the assessment of probe performance in biologically relevant regions.
Platform-specific coverage biases also significantly affect probe evaluation. Relative genome coverage with the UG 100 platform drops significantly in mid-to-high GC-rich regions compared to the NovaSeq X Series [15]. This lack of coverage in GC-rich regions could exclude genes with known disease associations from analysis and interpretation, potentially skewing performance metrics for probes targeting these regions [15]. Such platform characteristics must be considered when designing probe evaluation studies and interpreting resulting performance metrics.
The evaluation of probe performance through on-target rate, uniformity, and specificity provides critical insights for selecting and optimizing targeted sequencing approaches. As the field advances, methods like Linked Target Capture and improved amplicon-based approaches offer solutions to traditional limitations of hybridization-based enrichment. By applying standardized evaluation metrics and experimental protocols across platforms, researchers can make informed decisions that maximize sequencing efficiency and data quality for their specific applications. The continuing evolution of probe technologies promises even greater precision and efficiency in targeted sequencing, further enabling researchers to focus on genomically precise regions of interest with confidence and reliability.
Managing Computational and Data Storage Challenges
The choice between whole genome sequencing (WGS) and targeted sequencing represents a fundamental trade-off between genomic comprehensiveness and resource allocation. For researchers, scientists, and drug development professionals, this decision directly impacts computational infrastructure, data storage requirements, and analytical workflows. While WGS aims to capture the complete genetic blueprint, targeted sequencing focuses on specific genomic regions of interest, yielding significantly smaller, more manageable datasets. This guide objectively compares the performance and technical requirements of these approaches to inform strategic planning for genomics research.
Technology Comparison: Whole Genome vs. Targeted Sequencing
Table 1: Key Characteristics of Whole Genome and Targeted Sequencing Approaches
| Feature | Whole Genome Sequencing (WGS) | Targeted Sequencing |
|---|---|---|
| Genomic Coverage | Interrogates the entire genome, including coding (exons) and non-coding regions (introns) [1]. | Focuses on specific regions: individual genes, exomes (all protein-coding regions, ~2% of genome), or targeted panels [1]. |
| Primary Advantage | Provides a complete, hypothesis-free view of the genome, enabling discovery of novel variants outside known regions. | Enables much higher sequencing depth for lower cost, providing more confidence in detecting low-frequency variants [1]. |
| Typical Application | Discovery research, identification of novel biomarkers, comprehensive genetic studies. | Clinical diagnostics, validation studies, focused panels for actionable genes (e.g., in cancer) [1]. |
| Data Volume per Sample | Very high (typically tens to hundreds of gigabytes of raw data) [70]. | Significantly lower, proportional to the size of the targeted region. |
| Computational Load | High demands for data processing, alignment, and variant calling across billions of base pairs. | Reduced requirements for data processing and storage. |
Performance and Accuracy Benchmarking
The performance of sequencing platforms is critical for data integrity and impacts downstream storage and analysis. Benchmarking against standardized references, such as the Genome in a Bottle (GIAB) consortium benchmarks from the National Institute of Standards and Technology (NIST), is essential for evaluating platform accuracy [15].
Table 2: WGS Platform Performance Benchmarking Based on NIST v4.2.1 (HG002) [15]
| Metric | Illumina NovaSeq X Series | Ultima Genomics UG 100 Platform |
|---|---|---|
| Benchmark Region | Full NIST v4.2.1 benchmark | Subset ("High-Confidence Region") excluding 4.2% of the genome |
| SNV Errors | Baseline | 6× more errors |
| Indel Errors | Baseline | 22× more errors |
| Challenging Regions | Maintains high coverage and accuracy in GC-rich sequences and long homopolymers (>10 bp) | Decreased coverage in GC-rich regions; HCR excludes homopolymers longer than 12 bp |
| ClinVar Variants Excluded | 0% | 1.0% of variants excluded from analysis |
Independent comparative studies, even between older platforms, highlight that variant calling concordance is a persistent challenge. One study comparing Illumina and Complete Genomics technologies found that while 88.1% of single-nucleotide variants (SNVs) were concordant, there were tens of thousands of platform-specific calls, and only 26.5% of insertions and deletions (indels) were concordant [71]. This underscores the computational challenge of resolving discrepancies and the storage burden of maintaining raw data for re-analysis.
Experimental Protocols for Performance Validation
The following methodologies are representative of those used to generate the comparative data cited in this guide.
Protocol for Comparative Analysis of WGS Platforms
This protocol is based on Illumina's internal analysis comparing NovaSeq X Series to the Ultima Genomics UG 100 platform [15].
- Sample & Sequencing:
- Illumina Data: WGS data was generated on a NovaSeq X Plus System using the NovaSeq X Series 10B Reagent Kit. Secondary analysis was performed using DRAGEN v4.3. Data was downsampled to 35× coverage.
- Ultima Data: Publicly available WGS data generated on the UG 100 platform at 40× coverage was sourced, which had been analyzed using DeepVariant software by Ultima Genomics.
- Variant Calling & Benchmarking:
- Variant calling performance for both platforms was assessed against the full NIST v4.2.1 benchmark for the GIAB HG002 reference genome.
- The analysis specifically compared the number of false positives (variants called not in the benchmark) and false negatives (benchmark variants not called).
- Performance was also evaluated in challenging genomic regions, including GC-rich areas and homopolymers.
Protocol for Target Enrichment Sequencing Comparison
This protocol is adapted from a study comparing target enrichment methods for sequencing the Hantaan orthohantavirus genome, illustrating the considerations for targeted approaches [72].
- Sample Preparation: RNA was extracted from Apodemus agrarius lung tissues. Viral RNA copy number was quantified using reverse transcription quantitative PCR (RT-qPCR).
- Library Preparation & Enrichment: Three different enrichment methods were applied prior to sequencing on an Illumina MiSeq platform:
- Sequence-Independent, Single-Primer Amplification (SISPA): A method for random amplification of nucleic acids without prior targeting.
- Target Capture: Fragmented and adapter-ligated libraries were enriched using virus-specific probes.
- Amplicon NGS: A tiling scheme of primers was used to amplify short, overlapping fragments covering the entire viral genome.
- Analysis: The depth of coverage and breadth of coverage (percentage of the genome covered) for each method were analyzed and compared based on the initial viral RNA copy number.
Experimental Workflow and Decision Logic
The diagram below outlines the key decision points and workflows when choosing between whole genome and targeted sequencing strategies.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagent Solutions for Sequencing Workflows
| Item | Function in the Workflow |
|---|---|
| NovaSeq X Series 10B Reagent Kit (Illumina) [15] | Provides the chemistry (enzymes, nucleotides, buffers) for massive parallel sequencing on the NovaSeq X platform, determining data output and quality. |
| DRAGEN Secondary Analysis Platform (Illumina) [15] | A dedicated bioinformatics platform for secondary analysis (alignment, variant calling) that uses hardware acceleration to significantly reduce computation time and resource load. |
| Target Enrichment Kits (e.g., Agilent SureSelect) [71] | Kits containing probes or baits designed to capture specific genomic regions of interest from a complex DNA library prior to sequencing, enabling targeted sequencing. |
| Amplicon-Based Panel Kits [73] | Pre-designed sets of primers to amplify a specific set of genes or regions via multiplex PCR, used for creating targeted sequencing libraries. |
| Molecular Inversion Probe (MIP) Kits [73] | A type of probe used for targeted capture that can distinguish between very similar sequences, useful for SNP detection and copy number variant (CNV) analysis. |
| DeepVariant Software [15] | A deep learning-based variant calling tool that converts sequencing alignment data into called SNPs and indels, representing a modern computational approach. |
Strategic Considerations for Resource Management
The choice between WGS and targeted sequencing has direct and significant implications for managing computational and data storage resources.
- Infrastructure Investment: WGS demands robust, high-performance computing clusters and extensive storage arrays, often requiring petabyte-scale solutions for large cohorts. Targeted sequencing can often be performed with more modest on-premise servers or even through cloud-based analysis platforms.
- Cost Dynamics: While the per-base cost of sequencing has plummeted to as low as $350-$500 for a whole genome, the total cost of ownership must include data storage and analysis [24]. The Genomics Costing Tool (GCT), co-developed by organizations including FIND and the WHO, helps laboratories model these expenses, demonstrating that increased throughput can significantly reduce the cost per sample [60].
- Data Management: Effective data lifecycle policies are crucial. This includes defining protocols for how long raw data (FASTQ), processed alignment files (BAM), and final variant calls (VCF) are retained, and implementing data compression and archiving strategies to optimize storage utilization.
Addressing Interpretation Hurdles in Non-Coding and Complex Genomic Regions
The choice between whole genome sequencing (WGS) and targeted sequencing represents a fundamental strategic decision in genomic research, carrying significant implications for the interpretation of non-coding and complex genomic regions. WGS analyzes the complete DNA sequence of an organism, encompassing all coding and non-coding regions, typically to identify a comprehensive range of genetic aberrations including single nucleotide variants, insertions, deletions, and copy number variants [1]. In contrast, targeted sequencing focuses on a preselected subset of the genome, such as specific genes or coding regions known to harbor disease-relevant mutations, enabling much higher sequencing depth at a lower cost [1]. The central challenge in genomic interpretation lies in the fact that exomes—the protein-coding regions targeted in whole-exome sequencing (WES) and many panels—comprise a mere 2% of the human genome [1], leaving the vast landscape of non-coding DNA largely unexplored by targeted approaches.
The functional interpretation of non-coding regions presents substantial hurdles because regulatory elements have high evolutionary turnover, which obfuscates the use of conservation-based analysis methods for many genomic regions [74]. Furthermore, non-coding regions exhibit complex functional relationships where the same genetic variant can have divergent effects depending on its genomic context. Advanced methodologies are now required to decipher functionality in these regions, with intolerance to variation emerging as a strong predictor of human disease relevance independent of evolutionary conservation [74].
Technical Comparison of WGS and Targeted Sequencing
The technical and performance characteristics of WGS and targeted sequencing diverge significantly, influencing their applicability for different research scenarios, particularly those involving non-coding regions.
Table 1: Performance Characteristics of Sequencing Approaches
| Parameter | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Panels |
|---|---|---|---|
| Genomic Coverage | Comprehensive (3 billion base pairs) | ~2% of genome (exonic regions only) | Select genes/regions (often < 1%) |
| Sequencing Depth | Typically 30-50x for population studies | Typically 100-200x | Often >500x |
| Ability to Interrogate Non-Coding Regions | Complete access to promoters, enhancers, introns, intergenic regions | Limited to proximal non-coding regions (UTRs, splice sites) | Restricted to predefined non-coding targets (if included) |
| Variant Detection Spectrum | SNVs, indels, CNVs, structural variants, non-coding variants | Primarily coding SNVs and indels | Predesigned SNVs, indels, or fusions |
| Cost Considerations | Higher per sample | Moderate | Lower per sample |
| Informatics Complexity | High data storage and computational needs | Moderate | Lower |
Table 2: Applications in Non-Coding and Complex Region Analysis
| Analysis Type | WGS Performance | Targeted Sequencing Performance |
|---|---|---|
| Non-Coding Variant Discovery | Comprehensive detection of novel regulatory variants | Limited to predefined non-coding targets |
| Structural Variant Detection | Excellent for intergenic and intragenic SVs | Limited to targeted gene rearrangements |
| Epigenomic Correlation | Enables integration with methylation and chromatin data | Restricted to specific correlated sites |
| Haplotype Resolution | Phasing across entire loci and gene clusters | Limited phasing within targeted regions |
| Rare Variant Detection | Moderate in non-coding regions (due to lower depth) | Excellent for targeted hotspots |
Recent performance data from clinical research settings demonstrates that comprehensive genomic profiling using WGS approaches can simultaneously analyze hundreds of genes while capturing non-coding regulatory elements, whereas targeted panels like the 1,080-gene oncology panel provide ultra-deep coverage but limited genomic context [75]. The emerging trend shows that WGS consistently achieves lower failure rates compared to targeted sequencing or array-based platforms in applications like non-invasive prenatal testing, suggesting advantages in complex genomic regions [46].
Advanced Methodologies for Non-Coding Region Interpretation
Constraint-Based Prioritization with gwRVIS and JARVIS
The genome-wide residual variation intolerance score (gwRVIS) represents a breakthrough approach for identifying non-coding regions under evolutionary constraint. This method applies a sliding-window approach across whole genome sequencing data from 62,784 individuals to quantify intolerance to variation throughout the genome [74]. The resulting score identifies regions that are preferentially depleted of genetic variation due to purifying selection—an indicator of functional importance.
The computational workflow for gwRVIS begins with quality control and variant preprocessing from WGS data, followed by a sliding-window analysis (3kb windows with 1-nucleotide step) that records all variants and common variants (MAF > 0.1%) within each window [74]. An ordinary linear regression model predicts common variants based on the total number of all variants found in each window, with the studentized residuals of this regression defining the gwRVIS score, where lower values indicate greater intolerance to variation [74].
Building upon this foundation, JARVIS integrates gwRVIS with functional genomic annotations and primary genomic sequence using deep learning to create a comprehensive framework for prioritizing non-coding regions [74]. This approach intentionally excludes evolutionary conservation data, enabling the identification of human-lineage-specific constraint patterns that may be missed by conservation-based methods. When validated against known genomic elements, these methods successfully stratify functional classes by intolerance level, with ultraconserved noncoding elements (UCNEs) emerging as the most intolerant class (median gwRVIS: -0.99), followed by VISTA enhancers (-0.77) and protein-coding CCDS regions (-0.55) [74].
Regional Methylation Analysis with Principal Components
For interpreting epigenetic regulation in non-coding regions, the regionalpcs method addresses critical limitations in conventional DNA methylation analysis. This approach uses principal components analysis (PCA) to capture complex methylation patterns across gene regions, contrasting with traditional averaging methods that oversimplify correlation structures between CpG sites [76].
The experimental protocol for regionalpcs analysis involves:
- Data Acquisition: Whole-genome bisulfite sequencing or reduced representation bisulfite sequencing (RRBS) data
- Region Definition: Annotation of genomic regions (full genes, promoters, CpG islands, or custom regions)
- PCA Implementation: Decomposition of methylation variance across CpGs within each region
- Component Selection: Application of Gavish-Donoho method to identify optimal number of components
- Downstream Analysis: Association testing using regional principal components (rPCs) instead of individual CpGs
In simulation studies, this method demonstrated a 54% improvement in sensitivity over averaging approaches for detecting differentially methylated regions [76]. When 25% of CpGs were differentially methylated, rPCs detected a median of 73.1% of affected regions compared to just 19.1% with averaging. Performance advantages were particularly pronounced in scenarios with subtle methylation differences (1% difference: 18.8% vs 8.4% detection) and smaller sample sizes (50 samples: 94.4% vs 32.6% detection) [76].
Research Reagent Solutions for Non-Coding Genomic Studies
Table 3: Essential Research Reagents and Platforms
| Reagent/Platform | Function | Application in Non-Coding Studies |
|---|---|---|
| DNBSEQ-T1+ System [75] | High-throughput sequencing platform | Cost-effective WGS and exome studies for non-coding region analysis |
| DNBSEQ-G99RS Flow Cells [75] | Adjustable throughput sequencing | Flexible scaling for targeted panels and exome-scale testing |
| OmicsNest Bioinformatics Platform [75] | End-to-end analysis for microbial identification and assembly | Streamlines bioinformatics workflows for metagenomic and targeted sequencing |
| CRISPR-Based Enrichment Workflows [48] | Programmable target enrichment | Higher specificity in GC-rich or repetitive non-coding loci |
| Ultra-sensitive WGS-based ctDNA Monitoring [75] | Minimal residual disease detection | Non-coding variant tracking in liquid biopsies |
| Library Preparation Kits [6] | DNA fragment preparation for sequencing | Optimized for either WGS or targeted approaches |
| Target Enrichment Kits [6] | Probe-based capture of genomic regions | Selection of non-coding elements for focused studies |
Emerging Technologies and Future Directions
The landscape of non-coding region interpretation is rapidly evolving with several technological innovations poised to address current limitations. Third-generation sequencing platforms from Oxford Nanopore Technologies and Pacific Biosciences are expanding read lengths, enabling real-time, portable sequencing that improves resolution of complex genomic regions [77]. The recent Guinness World Record for fastest whole human genome sequencing at 3 hours 57 minutes demonstrates the accelerating pace of analytical workflows, bringing same-day genetic analysis closer to clinical reality [45].
Artificial intelligence and machine learning are increasingly critical for deciphering non-coding function. Tools like Google's DeepVariant utilize deep learning to identify genetic variants with greater accuracy than traditional methods [77]. AI models are also being applied to analyze polygenic risk scores and predict disease susceptibility by integrating coding and non-coding variants. The combination of AI with multi-omics data (transcriptomics, proteomics, metabolomics, epigenomics) provides a more comprehensive view of biological systems, linking non-coding genetic information with molecular function and phenotypic outcomes [77].
Market analysis indicates substantial growth in these sectors, with the whole genome and exome sequencing market projected to grow from $2.02 billion in 2024 to $6.14 billion in 2029 at a compound annual growth rate of 24.9% [6]. This expansion is fueled by population genomics initiatives, rising demand for precision medicine, and expanding applications in rare disease research—all areas where non-coding variant interpretation plays an increasingly important role [6].
Direct Performance Comparison and Validation Metrics
Within precision medicine, the choice of genomic sequencing approach is foundational. The debate between comprehensive Whole Genome Sequencing (WGS) and focused Targeted Sequencing Panels is central to research and diagnostic strategy. This guide provides an objective, data-driven comparison of these technologies, detailing their performance characteristics, optimal applications, and experimental protocols to inform decision-making by researchers, scientists, and drug development professionals.
At-a-Glance Comparison of Core Technologies
The table below summarizes the fundamental technical and operational differences between the main sequencing approaches.
| Feature | Whole Genome Sequencing (WGS) | Whole Exome Sequencing (WES) | Targeted Sequencing Panels |
|---|---|---|---|
| Genomic Coverage | Entire genome (~3 billion bases), including exons, introns, and non-coding regions [1]. | Protein-coding exons only (~2% of the genome, ~30-50 million bases) [1]. | Select genes or genomic regions known to harbor disease-associated mutations [1]. |
| Variant Types Detected | Broad range: SNVs, Indels, CNVs, SVs, repeat expansions, and variants in regulatory regions [78]. | Primarily SNVs and small Indels within exons; limited capacity for other variant types [1]. | Focused on pre-defined SNVs, Indels, and sometimes CNVs/fusions within the panel [1] [79]. |
| Sequencing Depth | Typically lower (e.g., 30x-40x) for standard coverage [79]. | High (often >100x) due to smaller target size [1]. | Very high (often 500x-1000x+), enabling detection of low-frequency variants [1] [79]. |
| Cost (Relative) | Higher | Moderate | Lower [1] |
| Best Application | Discovery of novel variants, complex disease research, comprehensive structural variant analysis, and as a universal first-tier test [78]. | Cost-effective alternative to WGS for identifying coding variants associated with Mendelian disorders [1]. | Clinical diagnostics for known conditions, somatic mutation profiling in oncology, and screening for specific, actionable biomarkers [1] [79]. |
Performance Benchmarking and Experimental Data
Diagnostic Yield and Variant Detection Sensitivity
Recent studies directly comparing these methodologies in clinical cohorts provide critical performance data. A key 2024 study compared a Target-Enhanced WGS (TE-WGS) approach against the TruSight Oncology 500 (TSO500) targeted panel in 49 patients with solid cancers [79]. The TE-WGS method, which combined a standard WGS backbone (40x coverage) with deep sequencing (500x) of over 500 key biomarker genes, demonstrated exceptional performance [79].
- Sensitivity: TE-WGS detected 100% (498/498) of the variants reported by the TSO500 panel [79].
- Variant Allele Fraction Correlation: A very high correlation (r=0.978) was observed between the variant allele fractions measured by both platforms, indicating high concordance in quantitative variant measurement [79].
- Added Value of WGS: Crucially, the matched normal (blood) TE-WGS data revealed that 44.8% (223/498) of the variants detected in the tumor were of germline origin, a distinction that is challenging with tumor-only targeted sequencing. The remaining 55.2% (275) were confirmed as bona fide somatic variants [79].
For mitochondrial DNA (mtDNA) analysis, a large-scale study of 1,499 individuals compared WGS with mtDNA-targeted sequencing. It found that both methods have comparable capacity for calling genotypes, haplogroups, and homoplasmies. However, significant variability was observed in calling low-frequency heteroplasmies, indicating that the detection of minor variant populations is highly method-dependent and requires cautious interpretation [5].
Platform-Specific Accuracy in Challenging Genomic Regions
Sequencing accuracy is not uniform across the genome. Repetitive sequences, homopolymers, and GC-rich regions pose significant challenges. A comparative analysis of the Illumina NovaSeq X Series and the Ultima Genomics UG 100 platform highlights these differences, which are relevant when selecting a platform for WGS [15].
- Variant Calling Errors: When assessed against the full NIST v4.2.1 benchmark, the UG 100 platform resulted in 6 times more single nucleotide variant (SNV) errors and 22 times more insertion/deletion (indel) errors than the NovaSeq X Series [15].
- Coverage in Challenging Regions: The UG 100 platform's "high-confidence region" (HCR) excludes 4.2% of the genome, including many challenging segments. In contrast, the NovaSeq X Series maintains high coverage and accuracy in these regions [15].
- Impact on Disease Genes: The NovaSeq X Series provided superior coverage and fewer indel errors in clinically critical, GC-rich genes like B3GALT6 (linked to Ehlers-Danlos syndrome) and the tumor suppressor BRCA1, where 1.2% of pathogenic variants fall within regions excluded by the UG 100 HCR [15].
Diagram 1: Simplified Workflow Comparison between WGS and Targeted Sequencing. WGS skips the target enrichment step, analyzing the entire genome uniformly. Targeted sequencing requires a hybridization step to capture specific genes of interest before sequencing.
Detailed Experimental Protocols
To ensure reproducibility and provide context for the data presented, this section outlines the key methodologies from the cited studies.
- 1. Sample Preparation: Extract DNA from both FFPE tumor tissue and matched normal peripheral blood.
- 2. Library Construction: Prepare sequencing libraries using the TruSeq Nano Library Prep Kits (Illumina).
- 3. Whole Genome Sequencing: Sequence libraries on an Illumina NovaSeq6000 to achieve an average of 40x coverage for tumor and 20x coverage for normal samples.
- 4. Target Enrichment:
- Design hybridization probes (e.g., xGen Custom Probes from IDT) for a bed file encompassing 526 genes (including all genes on the TSO500 panel).
- Re-hybridize and enrich the tumor DNA libraries using these probes.
- 5. Deep Targeted Sequencing: Sequence the enriched libraries on an Illumina NovaSeq6000 to achieve an average depth of >500x coverage over the targeted regions.
- 6. Bioinformatic Analysis:
- Alignment: Align sequences to the GRCh38 reference genome using BWA-MEM.
- Variant Calling:
- Small Variants: Use Strelka2 and Mutect2 for somatic calls; use HaplotypeCaller and Strelka2 for germline calls.
- Structural Variants: Use Manta for calling SVs.
- Copy Number & Purity: Use Sequenza to estimate tumor cell fraction and copy number profiles.
- Variant Prioritization: For hotspot mutations, apply a minimum VAF cutoff of 1% with supporting reads.
- 1. Sample Cohort: 1,499 participants from the Severe Asthma Research Program (SARP).
- 2. Whole Genome Sequencing:
- Library Prep: 500 ng DNA input using a PCR-free kit (Kappa Hyper).
- Sequencing: Illumina HiSeq X, 150 bp paired-end reads.
- 3. mtDNA-Targeted Sequencing:
- Enrichment: Digest nuclear DNA with Exonuclease V, then perform whole mitochondrial genome amplification using the REPLI-g mtDNA Kit (QIAGEN).
- Library Prep: 2 ng of enriched mtDNA with Nextera XT kit (Illumina).
- Sequencing: Illumina MiSeq, 151 bp paired-end reads.
- 4. Bioinformatics & Analysis:
- Alignment: Align raw reads from both methods to the revised Cambridge Reference Sequence (rCRS) using BWA.
- Variant Calling: Call mtDNA variants (heteroplasmies and homoplasmies) using MitoCaller, a likelihood-based method that accounts for sequencing error and mtDNA circularity.
- Genotype Definition:
- Homoplasmy: Alternative Allele Frequency (AAF) > 95%
- Heteroplasmy: AAF between 5% and 95%
- Reference: AAF < 5%
The Researcher's Toolkit: Essential Reagents & Materials
The table below lists key reagents and tools used in the featured experiments, crucial for replicating these sequencing workflows.
| Product / Solution | Function / Application | Example Use Case |
|---|---|---|
| TruSeq Nano Library Prep Kit (Illumina) | Preparation of sequencing-ready libraries from genomic DNA. | Used in the TE-WGS protocol for both WGS and target-enrichment library construction [79]. |
| xGen Custom Hybridization Probes (IDT) | Target-specific probes designed to capture and enrich genomic regions of interest. | Used to deeply sequence 526 key cancer genes in the TE-WGS study [79]. |
| REPLI-g Mitochondrial DNA Kit (QIAGEN) | Specifically amplifies the entire mitochondrial genome while minimizing nuclear DNA co-amplification. | Used for mtDNA enrichment in the targeted-seq protocol for the SARP cohort [5]. |
| Nextera XT DNA Library Prep Kit (Illumina) | Rapid preparation of sequencing libraries from low DNA input. | Used to prepare libraries from the amplified mtDNA in the targeted-seq protocol [5]. |
| DRAGEN Secondary Analysis (Illumina) | Integrated, hardware-accelerated bioinformatic platform for primary and secondary NGS analysis. | Used for variant calling and analysis in benchmarking studies of the NovaSeq X Series [15]. |
| MitoCaller Software | A specialized, likelihood-based variant caller for detecting heteroplasmy and homoplasmy in mtDNA. | Used to call mtDNA variants in the comparative study of WGS vs. targeted-seq [5]. |
Diagram 2: Sequencing Technology Selection Guide. A decision-flow diagram to help researchers select the most appropriate sequencing technology based on their primary objective and budget constraints.
The choice between Whole Genome Sequencing and Targeted Sequencing is not a matter of which technology is universally superior, but which is most fit-for-purpose. WGS stands out as a powerful, hypothesis-free discovery tool and a comprehensive clinical test, capable of identifying a wide range of variant types across the entire genome. Targeted panels offer a cost-effective, highly sensitive solution for focused applications where the genetic targets are well-defined, such as in routine oncology testing or for validating specific biomarkers.
Emerging methodologies like Target-Enhanced WGS demonstrate a powerful synergy, combining the breadth of WGS with the sensitivity of targeted sequencing. As sequencing costs continue to fall and bioinformatic tools advance, WGS is poised to become more accessible. However, the rigorous benchmarking of platforms and thoughtful consideration of clinical utility and workflow integration, as detailed in this guide, will remain essential for leveraging these technologies to their fullest potential in research and drug development.
Analyzing Concordance and Platform-Specific Variant Calls
Next-generation sequencing (NGS) has revolutionized genomic research and clinical diagnostics, offering multiple approaches for variant discovery. The two predominant strategies—whole genome sequencing (WGS) and targeted sequencing (TS)—each present distinct advantages and limitations in the critical assessment of concordance and platform-specific variant calls [1]. Concordance, defined as the consistency of variant detection across different sequencing platforms or methodologies, serves as a fundamental metric for establishing technical reliability in genomic applications. Platform-specific variant calls—discrepancies in mutation identification attributable to the sequencing technology itself—represent a significant challenge for clinical interpretation and research reproducibility [80] [81].
The broader thesis of WGS versus targeted sequencing research extends beyond mere technical comparisons to address foundational questions in genomic medicine: how to achieve optimal sensitivity and specificity across diverse genomic contexts, how to balance comprehensive coverage against practical constraints, and how to establish confidence in variant calling for clinical decision-making. This guide objectively compares the performance of these approaches through experimental data, methodological protocols, and analytical frameworks to inform researchers, scientists, and drug development professionals.
Foundational Sequencing Approaches and Their Performance Characteristics
Whole Genome Sequencing: Comprehensive but Complex
WGS aims to determine the order of all nucleotides (A, C, G, T) across an entire genome, capturing both coding and non-coding regions [1]. This comprehensiveness enables detection of variants throughout the genome, including intronic regions that may regulate gene expression [1]. PCR-free WGS protocols have demonstrated superior uniformity of coverage and minimal GC bias compared to other methods, achieving near-complete coverage of coding regions (100% of RefSeq exons in one study) [82]. This approach also facilitates robust copy number variation (CNV) detection and structural variant identification due to genome-wide coverage uniformity [82].
Targeted Sequencing: Depth-Focused and Efficient
Targeted sequencing concentrates on specific genomic regions of interest—typically genes with established disease associations—using enrichment techniques such as hybrid capture or amplicon-based approaches [12]. By focusing on a limited genomic footprint, TS achieves substantially higher sequencing depths (often exceeding 1000×) at lower cost and with reduced data burdens [83] [12]. This heightened depth enables reliable detection of low-frequency variants crucial for cancer research (somatic mutations) and liquid biopsy applications [12]. Targeted panels specifically designed for pharmacogenes have demonstrated excellent performance with depth-of-coverage ≥20× for at least 94% of target sequences [83].
Hybrid Capture vs. Amplicon-Based Enrichment
The two primary targeted enrichment methods—hybrid capture and amplicon-based approaches—exhibit different performance characteristics. Hybrid capture utilizes oligonucleotide probes to pull down regions of interest from fragmented DNA libraries, offering superior flexibility in target design and better coverage of complex genomic regions [17]. Amplicon sequencing employs polymerase chain reaction (PCR) with target-specific primers to amplify regions of interest, providing simpler workflows and lower DNA input requirements but potentially introducing amplification biases [17].
Table 1: Fundamental Comparison of WGS and Targeted Sequencing Approaches
| Characteristic | Whole Genome Sequencing | Targeted Sequencing |
|---|---|---|
| Genomic Coverage | Comprehensive (coding, non-coding, regulatory) | Limited to predefined regions of interest |
| Typical Sequencing Depth | 30-100× | 100-1000× (up to 5000× for ultra-deep applications) |
| Variant Detection Spectrum | SNVs, Indels, CNVs, SVs, non-coding variants | Primarily SNVs and Indels in targeted regions |
| Data Volume per Sample | High (~90-150 GB) | Moderate (1-10 GB, depending on panel size) |
| Optimal Applications | Novel variant discovery, structural variant detection, non-coding region analysis | High-confidence variant detection in known genes, low-frequency variant calling, clinical diagnostics |
Comparative Performance Metrics Across Platforms
Platform-Specific Variant Calling Performance
Recent evaluations of sequencing platforms reveal distinctive variant calling profiles. The Sikun 2000, a desktop NGS platform, demonstrated competitive performance in whole genome sequencing applications when compared to established Illumina platforms [80]. In a comprehensive assessment using five well-characterized human Genomes in a Bottle (GIAB) samples, the Sikun 2000 showed slightly higher SNP recall (97.24% vs. 97.02%) and precision (98.48% vs. 98.30%) compared to the NovaSeq 6000 [80]. However, its indel detection performance was moderately lower (83.08% vs. 87.08% recall compared to NovaSeq 6000) [80]. This pattern highlights the platform-specific strengths and weaknesses that researchers must consider when designing experiments.
The DNBSEQ-Tx platform has been optimized for whole-genome bisulfite sequencing (WGBS) applications, with two library construction methods (DNBPREBSseq and DNBSPLATseq) specifically developed for this platform [84]. The DNB_SPLATseq method demonstrated superior coverage uniformity, particularly in CpG island regions, and required less input DNA while being amenable to automated library construction [84]. Such platform-specific optimizations significantly impact data quality and experimental feasibility for specialized applications like epigenomics.
Concordance Across Sequencing Methodologies
Variant concordance between different sequencing approaches reveals methodological biases and limitations. A systematic comparison between targeted gene sequencing (TGS) and whole exome sequencing (WES) identified significant disparities in variant detection [81]. When analyzing the same endometrial cancer samples, a substantial number of variants were detected exclusively by one method or the other, with false positives and false negatives occurring in both approaches [81]. Using variants identified by both TGS and WES as a "high-confidence set" improved overall accuracy, suggesting that orthogonal verification enhances reliability for critical applications [81].
For noninvasive prenatal testing (NIPT), whole-genome sequencing technologies have demonstrated lower failure rates compared to targeted approaches, with simplified PCR-free workflows that reduce assay complexity and improve turnaround time [46]. The comprehensive view across the entire genome provided by WGS offers more informative results than targeted methods that analyze only limited regions of select chromosomes [46].
Table 2: Quantitative Performance Metrics Across Sequencing Platforms
| Platform/Method | SNV Recall | SNV Precision | Indel Recall | Indel Precision | Key Strengths |
|---|---|---|---|---|---|
| Sikun 2000 | 97.24% | 98.48% | 83.08% | 85.98% | High SNP accuracy, low duplication rate (1.93%) |
| NovaSeq 6000 | 97.02% | 98.30% | 87.08% | 85.80% | Robust indel detection, established platform |
| NovaSeq X | 96.84% | 98.02% | 86.74% | 84.68% | High base quality (Q30: 97.37%) |
| DNBSEQ-Tx (WGBS) | N/A | N/A | N/A | N/A | Cost-effective large-scale methylation studies |
| PCR-free WGS | N/A | N/A | N/A | N/A | Complete exome coverage, minimal GC bias |
| Targeted Panels | >99.9%* | >99.9%* | Variable | Variable | Ultra-deep sequencing, low-frequency variants |
*For established variants in targeted regions with adequate coverage [83]
Experimental Protocols for Concordance Assessment
Reference Material-Based Validation
The Genome in a Bottle (GIAB) reference materials developed by the National Institute of Standards and Technology (NIST) provide a robust framework for assessing sequencing platform performance and variant calling concordance [17]. These well-characterized DNA samples (including GM12878, and Ashkenazi Jewish and Chinese trios) come with high-confidence "truth sets" of small variant and homozygous reference calls, enabling systematic evaluation of assay performance [17].
Protocol: GIAB-Based Panel Validation
- DNA Sample Preparation: Obtain GIAB reference materials from the Coriell Institute (RM 8398, RM 8392, RM 8393) and quantify using fluorometric methods [17].
- Library Preparation: Perform library construction using both hybridization capture (e.g., Illumina TruSight Rapid Capture) and amplicon-based (e.g., Ion AmpliSeq) methods according to manufacturer protocols [17].
- Sequencing: Sequence libraries to appropriate depth (≥100× for targeted panels, ≥30× for WGS) using platforms of interest [17] [80].
- Variant Calling: Generate variant call format (VCF) files using standard bioinformatics pipelines for each platform [17].
- Performance Assessment: Compare query VCF files against GIAB high-confidence variants using GA4GH benchmarking tools on precisionFDA [17].
- Metric Calculation: Calculate sensitivity [TP/(TP+FN)], precision [TP/(TP+FP)], and false discovery rate across variant types and genomic contexts [17].
This approach enables standardized performance assessment across different platforms and enrichment methods, identifying systematic errors and platform-specific variant calling challenges [17].
Inter-Platform Concordance Evaluation
For laboratories validating sequencing results across multiple platforms, a replicated study design provides the most rigorous assessment of concordance.
Protocol: Inter-Platform Concordance Assessment
- Sample Selection: Utilize diverse DNA samples (e.g., cell lines, patient specimens) representing various genetic backgrounds [80] [81].
- Replicated Sequencing: Process identical DNA samples through different sequencing platforms (e.g., Sikun 2000, NovaSeq 6000, NovaSeq X) using consistent library preparation methods where feasible [80].
- Data Processing: Implement uniform bioinformatics pipelines for read alignment (e.g., BWA), duplicate marking, and variant calling (e.g., GATK HaplotypeCaller) across all datasets [80].
- Variant Comparison: Calculate Jaccard similarity indices to measure concordance between platforms for both SNVs and indels [80].
- Stratified Analysis: Assess performance differences by variant type, genomic context (GC-rich regions, repetitive elements), and functional category [17] [81].
- False Positive/Negative Investigation: Manually review alignment files at discordant positions using visualization tools (e.g., Golden Helix GenomeBrowse) to identify technical artifacts [17].
This protocol revealed that SNV concordance between Sikun 2000 and Illumina platforms (92.42%) was actually higher than the concordance between different Illumina platforms (92.06%), while indel concordance was more variable (65.22-70.62%) [80].
Diagram 1: Experimental workflow for platform concordance assessment
Essential Research Reagent Solutions
Successful concordance studies require carefully selected reagents and reference materials. The following table details essential solutions for rigorous sequencing comparisons.
Table 3: Essential Research Reagents for Sequencing Concordance Studies
| Reagent Category | Specific Examples | Function in Concordance Studies |
|---|---|---|
| Reference Materials | GIAB samples (GM12878, AJ trios) [17] | Provides ground truth for variant calling accuracy assessment |
| Targeted Enrichment Kits | TruSight Rapid Capture [17], AmpliSeq Inherited Disease Panel [17], NimbleGen SeqCap EZ [81] | Enables comparison of different enrichment technologies |
| Library Preparation Systems | Nextera Rapid Capture [83], Ion AmpliSeq Library Kit 2.0 [17] | Standardized library construction across platforms |
| Bisulfite Conversion Kits | EZ DNA Methylation-Gold kit [84] | Essential for methylation-specific concordance studies (WGBS) |
| Quality Control Tools | Qubit dsDNA HS Assay [84] [17], Bioanalyzer HS DNA chip [17] | Ensures input DNA quality and library preparation success |
| Validation Reagents | Sanger sequencing reagents [81], Digital PCR assays [12] | Orthogonal validation of discordant variant calls |
Technical and Biological Factors Influencing Concordance
Multiple technical factors contribute to variant calling discordance across platforms. GC-rich regions consistently demonstrate lower concordance due to capture biases in hybrid selection-based methods and sequencing artifacts in amplification-heavy protocols [82] [81]. One study found that while PCR-free WGS covered 100% of GC-rich first exons, WES covered only 93.60% of these challenging regions [82]. Library preparation methods significantly impact reproducibility, with PCR-free protocols demonstrating superior uniformity compared to amplification-based approaches [82] [80].
The specific variant type dramatically affects concordance rates. While SNVs generally show high inter-platform concordance (>92% in most comparisons), indels display substantially lower agreement (65-87%) due to alignment challenges and platform-specific error profiles [80]. Variant allele frequency also critically influences detection consistency, with low-frequency variants (<5%) showing markedly higher discordance rates, particularly in moderate-depth WGS compared to ultra-deep targeted sequencing [12] [81].
Bioinformatics Pipelines and Their Impact
Variant calling algorithms and parameters significantly contribute to platform-specific variant calls. Even with identical sequencing data, different bioinformatics pipelines can produce markedly different variant sets [17]. The GATK HaplotypeCaller, widely used for WGS data, employs local de novo assembly to resolve complex variants, while tools designed for targeted data may prioritize different analytical approaches [80].
Strategies for Discordance Resolution:
- Multi-Algorithm Consensus: Employ multiple variant calling algorithms and consider only concordant calls for high-confidence variant sets [81].
- Manual Review: Visualize alignment files at discordant positions to identify alignment artifacts, strand biases, or other technical issues [17].
- Orthogonal Validation: Utilize Sanger sequencing, digital PCR, or mass spectrometry for independent confirmation of clinically significant discordant variants [83] [81].
- Platform-Specific Filtering: Implement custom filtering strategies based on known error profiles of each sequencing platform [80].
Diagram 2: Analytical framework for resolving discordant variant calls
The comprehensive analysis of concordance and platform-specific variant calls reveals that both WGS and targeted sequencing play complementary but distinct roles in genomic research and clinical applications. WGS provides unparalleled comprehensiveness for novel variant discovery and structural variant detection, while targeted sequencing offers superior cost-effectiveness and sensitivity for established variant panels [82] [12].
For clinical applications requiring the highest possible accuracy, a tiered approach may be optimal: using targeted panels for established clinical variants where ultra-deep sequencing provides maximal sensitivity, while reserving WGS for complex cases where structural variants or novel mutations are suspected [82] [12]. In research settings, PCR-free WGS emerges as the most comprehensive approach for exploratory studies, while targeted sequencing remains ideal for large-scale cohort studies focusing on predefined genomic regions [82].
The consistent demonstration of platform-specific variant profiles underscores the importance of methodological transparency in publications and validation frameworks for clinical test development. As sequencing technologies continue to evolve, ongoing concordance assessments using standardized reference materials and protocols will remain essential for maintaining reproducibility and reliability in genomic science.
The choice between whole genome sequencing (WGS) and targeted sequencing (TS) represents a fundamental strategic decision in genomics research. While WGS aims to comprehensively sequence the entire genome, TS focuses on specific genes or regions of interest, enabling deeper coverage at a lower cost [12] [1]. Validating the results from either platform is crucial for ensuring data quality and reliability, forming an essential component of any rigorous sequencing workflow. This guide objectively compares the performance characteristics of WGS and TS, with a specific focus on the critical role of reference materials and public databases in the validation process, providing researchers with experimental data and methodologies to inform their sequencing strategies.
Technical Performance Comparison
Multiple studies have directly compared the analytical performance of WGS and TS approaches across different applications. The table below summarizes key performance metrics from recent comparative analyses:
Table 1: Performance Comparison of WGS and Targeted Sequencing
| Performance Metric | Whole Genome Sequencing | Targeted Sequencing | Comparative Experimental Findings |
|---|---|---|---|
| Sensitivity for SNVs/Indels | High for broad detection [85] | Very high for targeted regions [85] | TE-WGS demonstrated 96.3% sensitivity for variants identified by targeted panels in prostate cancer [85] |
| Coverage Uniformity | Genome-wide but can be variable [65] | Highly uniform across targeted regions [2] | Targeted sequencing achieves more consistent depth, critical for detecting low-frequency variants [12] |
| Variant Type Detection | Comprehensive (SNVs, Indels, CNVs, SVs) [65] [78] | Limited to panel design (SNVs, Indels, CNVs) [12] [2] | WGS identified an additional 430 clinically impactful variants (85%) missed by targeted panels [85] |
| Heteroplasmy Detection | Variable for low-frequency variants [5] | Comparable to WGS for homoplasmies/haplogroups [5] | Large variability in calling low-frequency heteroplasmies between methods; investigators should be cautious [5] |
| Structural Rearrangements | Excellent detection capability [78] [85] | Limited detection [2] | TE-WGS revealed rearrangements in BRCA1/2, RAD51B, NBN, and CDK12 missed by targeted panels [85] |
Experimental Protocols for Method Validation
Protocol 1: Cross-Platform Validation Study for Mitochondrial DNA
A 2021 study directly compared WGS and targeted-seq for analyzing mitochondrial DNA from 1,499 participants in the Severe Asthma Research Program, providing a robust framework for methodological validation [5].
Experimental Methodology:
- Sample Preparation: DNA was extracted from whole blood samples. WGS was performed at the New York Genome Center using the Kappa Hyper Library Preparation Kit (PCR-free) with 500 ng DNA input. Targeted sequencing was performed using nuclear DNA digestion, whole mitochondrial genome amplification with REPLI-g mitochondrial DNA kit, and Nextera XT DNA library preparation with 2 ng of mtDNA-enriched sample [5].
- Sequencing Platforms: WGS used Illumina HiSeq X with 150 bp paired-end reads. Targeted sequencing used Illumina MiSeq System with 151 bp paired-end reads [5].
- Bioinformatic Analysis: Raw sequencing data were aligned to the revised Cambridge Reference Sequence (rCRS) using BWA. Variants were called using MitoCaller, a likelihood-based method that accounts for sequencing error rate and mtDNA circularity [5].
- Validation Approach: The study compared genotype concordance, haplogroup determination, and heteroplasmy calling between platforms. Heteroplasmy was defined by alternative allele frequency between 5% and 95%, with homoplasmy above 95% [5].
Key Validation Findings: The study revealed that targeted-seq and WGS have comparable capacity to determine genotypes and call haplogroups and homoplasmies. However, significant variability was observed in calling heteroplasmies, particularly for low-frequency variants, highlighting the need for cautious interpretation of heteroplasmy data across different sequencing methods [5].
Protocol 2: Targeted-Enhanced WGS for Advanced Prostate Cancer
A 2024 study introduced Target-Enhanced Whole Genome Sequencing (TE-WGS) and compared it with clinical targeted panel sequencing (TPS) for advanced prostate cancer, demonstrating a novel approach to validation in oncology [85].
Experimental Methodology:
- Sample Cohort: 45 samples from 42 patients with metastatic prostate cancer were analyzed using both TE-WGS and TPS (Oncomine Comprehensive Panel, TruSight Oncology 500, or EXaCT-1) [85].
- WGS Protocol: Library preparation used Watchmaker DNA Library Preparation Kit with enzymatic fragmentation and adapter ligation. Sequencing was performed on Illumina NovaSeq 6000 with tumor samples at 40x coverage and matched germline at 20x coverage. Target-enhanced sequencing utilized xGen Custom Hybridization Probes targeting 2.76Mb at 500x depth [85].
- Variant Calling: Germline variants were called using HaplotypeCaller and Strelka2; somatic variants with Strelka2 and Mutect2; structural variants with Manta. All variants were annotated with Variant Effect Predictor and manually curated [85].
- Validation Metrics: Sensitivity was calculated for detecting TPS-reported variants, with additional analysis of clinically significant variants uniquely identified by each platform [85].
Key Validation Findings: TE-WGS demonstrated 96.3% sensitivity for detecting clinically relevant variants identified by TPS. Crucially, it identified additional actionable alterations in 46.7% of samples, including 35.6% with no actionable findings by TPS, highlighting the clinical value of comprehensive sequencing [85].
Essential Research Reagents and Databases
Validation of sequencing data requires both wet-lab reagents and bioinformatic resources. The table below outlines key solutions for rigorous experimental design:
Table 2: Research Reagent Solutions for Sequencing Validation
| Resource Type | Specific Examples | Function in Validation |
|---|---|---|
| Reference Materials | Genome in a Bottle (GIAB) Consortium [12], Genetic Testing Reference Materials Coordination Program (Get-RM) [12] | Provide characterized reference materials for assay development, quality control, validation, and proficiency testing |
| Variant Annotation | ANNOVAR [2], Variant Effect Predictor (VEP) [85] | Functional annotation of identified variants with population frequency, functional impact, and disease association data |
| Variant Calling | GATK [86] [2], MitoCaller [5], Mutect2 [85], Strelka2 [85] | Specialized algorithms for accurate identification of different variant types from sequencing data |
| Clinical Databases | ClinGen [12], ClinVar [2], Gen Curation Coalition (GenCC) [12] | Provide clinical interpretations of variants and gene-disease relationships for clinical reporting |
| Alignment Tools | BWA [5] [86] [85], Bowtie2 [86], BWA-MEM [85] | Map sequencing reads to reference genomes, forming the foundation for downstream variant calling |
| Phenotype Tools | Human Phenotype Ontology (HPO) [78], PhenoTips [78] | Standardize phenotypic data for correlation with genomic findings, improving diagnostic yield |
Sequencing Analysis Workflows
The bioinformatics workflows for WGS and targeted sequencing share common principles but differ in key aspects, particularly in the depth of analysis and data processing requirements. The following diagram illustrates the core steps and decision points in a standardized sequencing validation workflow:
Discussion and Best Practices
The experimental data presented demonstrates that both WGS and TS have distinct advantages depending on the research context. WGS provides unparalleled comprehensive variant detection, particularly for structural variants and rearrangements in non-coding regions, while TS offers superior depth for analyzing specific genomic regions of interest, often at a lower cost and with simpler data management [5] [12] [85].
For validation in practice, several best practices emerge:
- Platform Selection: Choose WGS for discovery-phase research or when analyzing genetically heterogeneous conditions without clear gene candidates. Opt for TS when focusing on well-characterized genes or when maximum depth for low-frequency variant detection is required [12] [1] [85].
- Reference Material Utilization: Incorporate characterized reference materials from GIAB or Get-RM throughout the workflow, from assay development to ongoing quality control, to ensure analytical validity [12].
- Database Integration: Leverage multiple public databases (ClinGen, ClinVar, GenCC) for clinical interpretation and avoid over-reliance on any single source to minimize interpretation biases [78] [12].
- Validation Design: When comparing platforms, utilize paired samples processed through both methods, as demonstrated in the mtDNA and prostate cancer studies, to directly assess analytical performance [5] [85].
As sequencing technologies continue to evolve, validation practices must similarly advance. The integration of reference materials and comprehensive database resources remains fundamental to ensuring the reliability and reproducibility of genomic findings, regardless of the sequencing platform employed.
Next-generation sequencing (NGS) has revolutionized biomedical research and clinical diagnostics, offering powerful tools for unraveling genetic contributions to disease. The two predominant approaches—whole genome sequencing (WGS) and targeted sequencing—each offer distinct advantages and limitations that make them suitable for different research scenarios. WGS provides a comprehensive view of the entire genome, including both coding and non-coding regions, enabling discovery of novel genetic elements across all 3 billion base pairs of the human genome. In contrast, targeted sequencing focuses on specific regions of interest, such as known disease-associated genes or pathways, allowing for deeper coverage at lower cost while generating more manageable datasets. Understanding the technical specifications, performance characteristics, and appropriate applications of each approach is essential for researchers designing genomic studies in cancer research, rare disease diagnosis, and pathogen surveillance.
This guide provides an objective comparison of WGS and targeted sequencing methodologies, supported by experimental data and performance metrics from recent studies. We examine the strengths and limitations of each approach across key application areas, provide detailed experimental protocols, and offer practical guidance for technology selection based on research objectives.
Technical Comparison of Sequencing Approaches
Fundamental Differences in Genomic Coverage
The primary distinction between WGS and targeted sequencing lies in the extent of genomic coverage. WGS sequences the entire genome, including exons, introns, intergenic regions, and structural elements, enabling comprehensive variant discovery across all genomic contexts. This approach is particularly valuable for identifying novel disease-associated variants in non-coding regulatory regions, structural rearrangements, and complex genomic alterations that may be missed by targeted approaches. Research demonstrates that non-coding regions spanning 98% of the human genome contain important regulatory elements, and somatic structural variants in cancer genomes remain widely unexplored without WGS approaches [87].
Targeted sequencing, including whole exome sequencing (WES) and gene panels, focuses on specific genomic regions of interest. WES targets the exome (approximately 2% of the genome) which contains ~85% of known disease-associated variants, while targeted panels sequence even smaller gene sets known to be associated with specific diseases [88] [89]. This focused approach allows for significantly higher sequencing depth (often 100-1000x) compared to typical WGS coverage (30-50x), enhancing sensitivity for detecting low-frequency variants. For clinical applications where speed, cost, and analytical simplicity are prioritized, targeted sequencing provides a practical solution for interrogating known disease-associated regions with high confidence [12] [1].
Performance Characteristics and Experimental Data
Recent benchmark studies have systematically evaluated the performance of WGS and targeted sequencing approaches across multiple platforms and laboratory sites. The SEQC2 consortium conducted a comprehensive cross-platform study using well-characterized reference samples to quantify accuracy, reproducibility, and factors affecting mutation detection. Their findings reveal distinct performance characteristics for each approach, summarized in the table below:
Table 1: Performance comparison of WGS and targeted sequencing approaches
| Parameter | Whole Genome Sequencing | Whole Exome Sequencing | Targeted Panels |
|---|---|---|---|
| Genome coverage | ~99% of entire genome [87] | ~2% (protein-coding regions) [1] | <1% (specific genes/regions) |
| Typical sequencing depth | 30-50x [87] | 100x [87] | 500-1000x or higher [12] |
| Variant detection sensitivity | High for novel variants [87] | Limited to coding regions [89] | Excellent for known targets [90] |
| Ability to detect structural variants | Comprehensive [87] | Limited [87] | Very limited |
| Data volume per sample | 90-150 GB [87] | 5-10 GB [87] | 0.5-1 GB [87] |
| Inter-center reproducibility | High [91] | Moderate with more batch effects [91] | Variable |
| Mutation calling consistency | High for SNVs, moderate for indels [87] | Affected by capture efficiency [91] | Highest for targeted regions |
The SEQC2 consortium study demonstrated that WES had better coverage-to-cost ratio than WGS but showed more batch effects and artifacts due to laboratory processing, resulting in larger variation between runs and laboratories [91]. WES also exhibited less reproducible results compared to WGS, particularly across different sequencing centers. The study also found that biological replicates were more important than bioinformatics replicates for achieving high specificity and sensitivity in mutation detection [91].
Application-Specific Case Studies
Cancer Genomics
In cancer research, the choice between WGS and targeted sequencing depends on the specific research questions, sample types, and available resources. WGS provides the most comprehensive mutation profiling, enabling detection of coding mutations, non-coding regulatory alterations, structural variants, and copy number changes across the entire genome. A study highlighting the utility of WGS in cancer research demonstrated its ability to identify novel structural variants and mutations in non-coding regions that may drive oncogenesis [87]. This comprehensive approach is particularly valuable for cancer types with complex genomic architectures or for discovery-oriented research aimed at identifying novel biomarkers.
Targeted sequencing panels have proven highly effective in clinical oncology for profiling known cancer-associated genes with high sensitivity, especially in samples with limited tumor content or low-quality DNA. These panels can detect variant allele frequencies as low as 0.1-0.2% in circulating tumor DNA (ctDNA), enabling applications in minimal residual disease monitoring [12]. A study by Frampton et al. demonstrated that a targeted cancer panel identified clinically actionable mutations in 76% of 2,221 tumors studied, significantly expanding therapeutic options compared to conventional diagnostic tests [12]. The focused nature of targeted panels makes them particularly suitable for clinical applications where specific therapeutic decisions rely on comprehensive mutation profiling of known cancer genes.
Table 2: Cancer genomics application case study comparison
| Characteristic | WGS Application | Targeted Sequencing Application |
|---|---|---|
| Research objective | Comprehensive driver mutation discovery [87] | Clinically actionable mutation profiling [12] |
| Sample type | High-quality tumor-normal pairs | FFPE, ctDNA, low-input samples [12] |
| Variant types detected | SNVs, indels, CNAs, SVs, non-coding [87] | SNVs, indels, focused gene regions [12] |
| Detection sensitivity | Moderate (limited by 30-50x depth) | High (500-1000x depth) [12] |
| Clinical actionability | Emerging, primarily research | High for known biomarkers [12] |
| Cost considerations | Higher per sample | Lower per sample, higher for large genes sets |
Rare Disease Diagnosis
In rare genetic disorders, WES has become the primary diagnostic approach due to its ability to interrogate all protein-coding regions where ~85% of disease-causing mutations reside [88] [89]. The unbiased nature of WES eliminates the need for preliminary candidate gene selection, making it particularly valuable for genetically heterogeneous conditions. Studies have demonstrated the success of WES in identifying novel Mendelian disease genes, with nearly 2,000 new entries added to OMIM since 2008 [89]. The focused nature of WES provides sufficient depth for reliable variant detection while maintaining reasonable costs and data management requirements.
For genetically heterogeneous rare diseases, targeted sequencing panels offer an efficient approach for analyzing known disease-associated genes. The TruSight One Sequencing Panel, for example, provides comprehensive coverage of >4,800 disease-associated genes, while the TruSight One Expanded Panel targets ~1,900 additional genes with recent disease associations [88]. These panels enable laboratories to focus resources on genes with established disease relationships, streamlining analysis and interpretation. For conditions like cystic fibrosis, targeted panels can provide comprehensive variant detection across diverse ethnic populations, overcoming the limitations of ethnicity-specific testing [88].
WGS is increasingly applied in rare disease diagnosis when WES is inconclusive, as it enables detection of non-coding and structural variants that may be disease-causing. While more expensive, WGS can identify pathogenic variants in regulatory regions, deep intronic mutations affecting splicing, and complex structural rearrangements missed by exome-based approaches [88].
Pathogen Surveillance and Infectious Disease
The COVID-19 pandemic highlighted the distinct utilities of WGS and targeted sequencing in pathogen surveillance and outbreak management. WGS provides complete genomic information for novel pathogen discovery, tracking transmission dynamics, and monitoring evolutionary trajectories. During the pandemic, WGS enabled researchers to understand SARS-CoV-2 transmission patterns, identify emerging variants of concern, and investigate the molecular basis of increased transmissibility or immune evasion [12].
Targeted sequencing approaches offer cost-effective solutions for high-throughput screening and specific variant detection. Amplicon-based panels focused on key viral genomic regions enabled efficient sequencing of thousands of SARS-CoV-2 samples, facilitating real-time surveillance with quick turnaround times [12]. These targeted approaches are particularly valuable in clinical settings where specific variant information guides treatment decisions or public health interventions.
In noninvasive prenatal testing (NIPT), WGS-based approaches demonstrate lower failure rates and simplified workflows compared to targeted methods [46]. The PCR-free sample preparation used in WGS-based NIPT reduces assay complexity and improves turnaround time, while providing comprehensive genomic coverage [46]. Targeted NIPT approaches, including SNP-based analysis and microarray methods, focus on specific chromosomal regions but require additional amplification steps that complicate workflows [46].
Experimental Design and Methodologies
Detailed Workflow Protocols
WGS Experimental Protocol: The standard WGS workflow begins with quality control of genomic DNA, followed by library preparation using either PCR-based or PCR-free protocols. For the TruSeq DNA PCR-Free protocol described in the SEQC2 consortium study [91], 1μg of input DNA is fragmented to approximately 350bp using Covaris sonication. Fragmented DNA undergoes end-repair, A-tailing, and adapter ligation before cleanup and quantification using fluorometry (Qubit or GloMax) and quality assessment by capillary electrophoresis (Bioanalyzer or TapeStation). Libraries are sequenced on platforms such as Illumina NovaSeq or HiSeq with 2×150bp reads, achieving 30-50x coverage. The PCR-free protocol reduces GC bias and provides more comprehensive coverage compared to PCR-based methods [87].
Targeted Sequencing Experimental Protocol: Targeted sequencing employs either amplicon-based or hybrid capture-based enrichment. The Illumina DNA Prep with Enrichment protocol [90] uses hybridization capture with custom or fixed panels to enrich for regions of interest. Library preparation begins with tagmentation of input DNA, followed by adapter ligation and PCR amplification. Libraries are hybridized with biotinylated probes targeting specific genomic regions, captured using streptavidin beads, and amplified before sequencing. This approach enables deep sequencing (500-1000x) of targeted regions while minimizing off-target coverage.
Bioinformatics Analysis Pipelines
WGS Analysis Pipeline: Cancer WGS analysis requires sophisticated computational pipelines to handle the large data volumes (approximately 1TB for tumor-normal pairs). The standard workflow begins with quality control of raw sequencing data (FASTQ files) using tools like FastQC. Reads are aligned to the reference genome (hg19 or hg38) using aligners such as BWAmem, followed by duplicate marking and base quality recalibration. Somatic mutation calling employs multiple algorithms specific to different variant types: MuTect2 for SNVs, Strelka for indels, Control-FREEC for CNAs, and Manta for SVs [87]. The ICGC benchmark study revealed that somatic indel calling shows high inconsistency across pipelines, while SNV and SV calls demonstrate better consensus [87].
Targeted Sequencing Analysis Pipeline: Analysis of targeted sequencing data follows similar principles but with focus on targeted regions. The DRAGEN Enrichment App provides an end-to-end solution for targeted panel data, including alignment, duplicate marking, and variant calling [90]. Enhanced depth of coverage in targeted regions enables more sensitive detection of low-frequency variants, with specialized tools like DRAGEN Somatic providing sensitive detection of low-frequency alleles in ctDNA applications [90].
Diagram 1: Technology selection framework for sequencing approaches
Essential Research Reagents and Platforms
The selection of appropriate reagents and platforms is critical for successful sequencing studies. The following table outlines key solutions for WGS and targeted sequencing workflows:
Table 3: Essential research reagents and platforms for sequencing studies
| Category | Product/Platform | Specifications | Applications |
|---|---|---|---|
| Library Prep | Illumina DNA Prep [90] | PCR-free or with PCR; 1-250ng input | WGS, WES, targeted panels |
| Library Prep | Illumina Cell-Free DNA Prep with Enrichment [90] | Specialized for low-input cfDNA | Liquid biopsy, ctDNA analysis |
| Enrichment | Illumina Exome 2.0 Plus Enrichment [88] | Comprehensive exome coverage | Whole exome sequencing |
| Enrichment | Illumina Custom Enrichment Panel v2 [90] | Custom target content | Targeted sequencing |
| Sequencing Systems | NovaSeq X Series [90] | Up to 16Tb output; 26B reads/flowcell | Large-scale WGS, population studies |
| Sequencing Systems | NextSeq 1000/2000 Systems [90] | Mid-throughput; fast turnaround | Targeted panels, exome sequencing |
| Bioinformatics | DRAGEN Bio-IT Platform [88] [90] | Hardware-accelerated analysis | Secondary analysis for all NGS types |
| Bioinformatics | DRAGEN Somatic [90] | Sensitive low-frequency variant detection | Cancer genomics, liquid biopsy |
The choice between WGS and targeted sequencing involves careful consideration of research objectives, sample characteristics, and resource constraints. WGS provides the most comprehensive approach for discovery-oriented research, enabling identification of novel variants across the entire genome. Its ability to detect structural variants, non-coding mutations, and complex genomic alterations makes it invaluable for advancing our understanding of disease genetics. However, the higher costs, substantial data management requirements, and analytical complexities present challenges for large-scale studies or routine clinical applications.
Targeted sequencing offers a practical solution for focused research questions and clinical applications where specific genes or regions are of interest. The enhanced sequencing depth achievable with targeted approaches provides superior sensitivity for detecting low-frequency variants in heterogeneous samples or liquid biopsies. The reduced data volumes and simplified analysis pipelines make targeted sequencing more accessible for laboratories with limited computational resources.
Future developments in sequencing technologies, including long-read sequencing, single-cell approaches, and integrated multi-omics, will further expand applications in biomedical research. The continuing reduction in sequencing costs will make WGS more accessible for routine applications, while improved target enrichment technologies will enhance the performance of targeted approaches. Regardless of technological advances, the fundamental trade-offs between comprehensiveness and depth will continue to inform selection of the appropriate sequencing strategy for specific research questions.
The choice between whole genome sequencing (WGS) and targeted sequencing represents a fundamental strategic decision for modern laboratories, balancing comprehensiveness against resource constraints. While whole genome sequencing examines the complete DNA makeup of an organism by determining the order of all nucleotides (A, C, G, T) across the entire genome, targeted sequencing focuses on a preselected subset of genes or genomic regions known to harbor mutations relevant to specific diseases [1]. This methodological distinction creates significant differences in the scale of data generated, computational resources required, and subsequent analytical complexity. As sequencing technologies advance and costs decrease—with WGS costs falling from approximately $100 million in 2001 to just over $500 in 2023 in the United States—the accessibility of these technologies has increased, making practical assessments of their computational burdens increasingly critical for laboratory planning and resource allocation [24].
Technical Comparison: Data Generation and Storage
The core difference between WGS and targeted sequencing lies in the sheer volume of data produced, which directly impacts storage requirements, computational processing time, and bioinformatic infrastructure needs.
Table 1: Direct Comparison of Data Generation and Computational Load
| Parameter | Whole Genome Sequencing (WGS) | Targeted Sequencing Panels | Whole Exome Sequencing (WES) |
|---|---|---|---|
| Genomic Coverage | Entire genome (~3.2 billion bases) | Selected genes/regions (variable size) | Protein-coding exons (~2% of genome) [1] |
| Data Volume per Sample | ~100 GB raw data [78] | Significanty lower (dependent on panel size) | ~5-10 GB raw data |
| Sequencing Depth | Typically 30-40x for standard analysis [79] | Often >500x for high-confidence variant calling [1] [79] | Typically 50-100x for reliable calling |
| Primary Data Burden | Extremely high | Low to moderate | Moderate |
| Processed Data Size | ~30 GB (CRAM/BAM/VCF) [78] | <1 GB (BAM/VCF) | ~3-5 GB (CRAM/BAM/VCF) |
Key Differentiators in Data Burden
Comprehensiveness vs. Efficiency: WGS provides a complete dataset that allows detection of a broad range of variant types—including single nucleotide variants (SNVs), insertions/deletions (indels), copy number variants (CNVs), structural variants (SVs), and repeat expansions—in a single assay without prior hypothesis about disease causation [78]. This comes at the cost of generating vast amounts of data, most of which resides in non-coding regions whose clinical significance may not yet be fully understood. In contrast, targeted sequencing generates focused datasets, enabling ultra-deep sequencing (500x or higher) of clinically actionable regions, which provides high confidence for detecting low-frequency variants but offers no information about regions outside the targeted panel [1] [79].
Storage Infrastructure Implications: The data volume from WGS has substantial infrastructure implications. A large-scale WGS project sequencing 1,000 genomes would generate approximately 100 terabytes of raw data, requiring significant and costly data storage solutions [78]. Targeted sequencing projects of similar scale produce orders of magnitude less data, making them more manageable for laboratories with limited computational infrastructure.
Experimental Approaches and Workflows
Understanding the data burden requires examination of the distinct experimental protocols and analytical workflows for each sequencing method. The following diagram illustrates the key differences in their data processing workflows and consequent computational demands.
Experimental Protocols and Data Generation
The experimental methodologies for WGS and targeted sequencing differ significantly in their initial steps, which directly influences subsequent data processing requirements.
WGS Laboratory Protocol: Standard WGS protocols, such as those referenced in the Medical Genome Initiative best practices, involve extracting DNA from samples (500 ng input typical), followed by PCR-free library preparation using kits such as the Kapa Hyper Library Preparation Kit [78]. Sequencing is then performed on high-throughput platforms like Illumina NovaSeq6000 or HiSeq X with paired-end reads (150 bp), generating approximately 100 GB of raw data per sample at 30-40x coverage [79] [78]. This approach avoids amplification biases but produces the maximum possible data volume from the sequencing platform.
Targeted Sequencing Laboratory Protocol: Targeted approaches begin with enrichment of specific genomic regions before sequencing. Methods include:
- Amplification-based approaches: Using long-range PCR to isolate mitochondrial DNA or other targets, as demonstrated in a study comparing WGS and targeted sequencing for mitochondrial DNA analysis [5].
- Hybridization capture: Employing customized probe sets (e.g., xGen Custom Hybridization Probes) to enrich for regions of interest, such as the 526-gene panel used in Target-Enhanced WGS (TE-WGS) methodology [79]. These enrichment techniques typically require only 20 ng of input DNA and generate dramatically less raw data while achieving much higher sequencing depth (500-1000x) in targeted regions [79] [5].
Bioinformatics Processing Pipelines
The secondary analysis—converting raw sequencing data to variant calls—represents another point of significant computational divergence.
WGS Analysis Workflow: The bioinformatic processing of WGS data demands substantial computational resources and involves:
- Alignment: Using tools like BWA-MEM to align reads to the reference genome (GRCh38), processing ~100 GB of FASTQ data per sample [79] [78].
- Variant Calling: Employing multiple specialized callers—Strelka2 for small variants, Manta for structural variants, Mutect2 for somatic mutations—each requiring significant processing time and memory [79].
- Annotation and Filtering: Adding functional context to millions of variants using tools like VEP, followed by phenotype-driven prioritization [78].
Targeted Analysis Workflow: The focused nature of targeted sequencing data enables more streamlined analysis:
- Alignment: Faster alignment processes due to significantly smaller dataset size.
- Variant Calling: Deep variant calling in targeted regions using tools like HaplotypeCaller or VarScan, with less computational intensity [5].
- Annotation: Limited to dozens to hundreds of variants, dramatically reducing the interpretation burden.
The Scientist's Toolkit: Essential Research Reagents and Computational Solutions
Successful implementation of either sequencing approach requires specific laboratory and computational resources. The table below details essential components for establishing these capabilities in a research setting.
Table 2: Research Reagent Solutions and Essential Materials for Sequencing workflows
| Category | Specific Products/Tools | Function in Workflow |
|---|---|---|
| Library Prep Kits | Kapa Hyper Library Preparation Kit (PCR-free) [78], Nextera XT DNA Library Prep Kit [5], TruSeq Nano Library Prep Kits [79] | Prepare DNA fragments for sequencing by adding adapters and indexes |
| Target Enrichment | xGen Custom Hybridization Probes [79], REPLI-g mitochondrial DNA kit [5] | Isolate and amplify specific genomic regions of interest for targeted sequencing |
| Sequencing Platforms | Illumina NovaSeq6000 [79], Illumina HiSeq X [78], Illumina MiSeq [5] | Generate raw sequencing data through massively parallel sequencing |
| Alignment Tools | BWA-MEM [79] [78] [5] | Map raw sequencing reads to reference genome |
| Variant Callers | Strelka2 [79], Mutect2 [79], Manta [79], MitoCaller [5] | Identify genetic variants from aligned sequencing data |
| Analysis Suites | DRAGEN Platform [15], CancerVision [79] | Integrated secondary analysis solutions for processing sequencing data |
| Data Storage | High-performance computing clusters, Cloud storage solutions | Store and manage large volumes of raw and processed sequencing data |
Performance Benchmarking: Experimental Data and Outcomes
Direct comparisons between WGS and targeted sequencing demonstrate their relative performance characteristics and analytical strengths.
Diagnostic Performance and Technical Validation
Recent studies have provided empirical data comparing the analytical performance of these approaches:
Oncology Application: A 2024 study comparing Target-Enhanced WGS (TE-WGS) with the targeted TruSight Oncology 500 (TSO500) panel demonstrated that TE-WGS detected all 498 variants identified by TSO500 (100% concordance) with a high correlation in variant allele fractions (r=0.978) [79]. Notably, TE-WGS provided additional clinical value by distinguishing germline from somatic variants through matched normal sequencing and delivered accurate copy number profiles, fusion genes, and genomic instability markers essential for comprehensive cancer management.
Mitochondrial DNA Analysis: A direct comparison of WGS and mtDNA-targeted sequencing using 1,499 paired samples revealed that both methods had comparable capacity for determining genotypes, calling haplogroups, and identifying homoplasmies [5]. However, significant variability emerged in detecting heteroplasmies, particularly low-frequency variants, highlighting methodological influences on specific variant types.
Platform-Specific Performance Considerations
Sequencing platform choice introduces additional variables affecting data burden and quality:
- Illumina NovaSeq X Series: Demonstrates high accuracy across challenging genomic regions, with variant calling accuracy of 99.94% for SNVs and 97% for CNVs when using DRAGEN secondary analysis, without excluding difficult-to-sequence regions [15].
- Ultima Genomics UG 100 Platform: Uses a "high-confidence region" (HCR) that excludes 4.2% of the genome—including homopolymer regions longer than 12 base pairs and certain GC-rich areas—which reduces computational burden but potentially misses biologically relevant variants in excluded regions [15].
Table 3: Comparative Experimental Results from Key Studies
| Study Metrics | Target-Enhanced WGS (TE-WGS) | TruSight Oncology 500 (Targeted) | Standard WGS | mtDNA-Targeted Sequencing |
|---|---|---|---|---|
| Variant Detection Sensitivity | 100% for TSO500 variants [79] | Benchmark for comparison | N/A | Comparable for homoplasmies [5] |
| Additional Findings | 44.8% variants of germline origin [79] | Limited to panel content | N/A | Variable for heteroplasmies [5] |
| Sequencing Depth | 40x WGS + 500x for targets [79] | ~500x [79] | 30-40x [78] | >1000x [5] |
| Data Volume | Higher than targeted, lower than standard WGS | Low | Very High (~100 GB) [78] | Very Low |
| Computational Load | High (combined analysis) | Moderate | Very High | Low |
The choice between WGS and targeted sequencing represents a strategic trade-off between comprehensiveness and resource efficiency. Whole genome sequencing provides the most complete genetic assessment but demands substantial computational infrastructure, data storage solutions, and bioinformatic expertise—with data burdens of approximately 100 GB per sample before analysis [78]. Targeted sequencing offers a resource-efficient alternative for focused research questions or clinical applications where established gene-disease relationships are well characterized, with significantly reduced data burdens and computational requirements.
Emerging hybrid approaches like Target-Enhanced WGS attempt to bridge this divide by combining the comprehensive backbone of WGS with deep sequencing of clinically relevant targets, though this approach still maintains a substantial data footprint [79]. Laboratories must weigh these technical considerations against their specific research objectives, clinical applications, and available computational resources when selecting the optimal sequencing strategy. As sequencing costs continue to decline and analytical methods improve, the field continues to evolve toward more efficient utilization of the vast data generated by comprehensive genomic approaches.
Conclusion
The choice between Whole Genome Sequencing and Targeted Sequencing is not a matter of superiority, but of strategic alignment with project objectives. WGS offers an unparalleled, comprehensive view of the genome, making it indispensable for novel discovery and complex disease research. In contrast, Targeted Sequencing provides a cost-effective, deep, and focused analysis ideal for routine clinical applications where speed, cost, and high sensitivity for known variants are paramount. The dramatic reduction in sequencing costs, with WGS now available for just over $500, is making comprehensive genomic analysis more accessible than ever. Future directions will see these technologies further integrated into personalized medicine, with WGS potentially becoming the first-line tool for diagnosis as interpretation frameworks mature. For drug development professionals, both methods are crucial for identifying and validating genetic targets, ultimately accelerating the creation of precision therapies. The key to success lies in a nuanced understanding of each method's strengths and a clear definition of the scientific or clinical question at hand.
References
- 1. - Whole . Genome -Exome vs . Whole ... Sequencing vs Targeted [https://www.healio.com/hematology-oncology/learn-genomics/whole-genome-sequencing/whole-genome-whole-exome-sequencing-and-targeted-sequencing-panels]
- 2. WGS vs. WES vs. Targeted Sequencing Panels - CD Genomics [https://www.cd-genomics.com/resource-wgs-vs-wes-vs-targeted-panels.html]
- 3. Panels, WES, or WGS: Which is Best for Rare Disease Diagnosis? [https://3billion.io/blog/which-is-the-best-ngs-approach-for-rare-disease-diagnosis-panels-wes-or-wgs]
- 4. Translational and clinical comparison of whole genome and transcriptome to panel sequencing in precision oncology | npj Precision Oncology [https://www.nature.com/articles/s41698-024-00788-3]
- 5. Comparison of whole genome sequencing and targeted sequencing for Mitochondrial DNA - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8354572/]
- 6. and Exome Whole Market Report Genome Sequencing 2025 [https://www.researchandmarkets.com/reports/5951784/whole-genome-exome-sequencing-market-report]
- 7. Global Targeted DNA Sequencing Market Innovation Report... [https://www.linkedin.com/pulse/global-targeted-dna-sequencing-market-innovation-aqj2e]
- 8. Decoding Sequencing Depth and Coverage - CD Genomics [https://www.cd-genomics.com/resource-sequencing-depth-and-coverage.html]
- 9. vs. Sequencing : Key Metrics in NGS Explained Depth Coverage [https://3billion.io/blog/sequencing-depth-vs-coverage]
- 10. How does whole differ from genome ... sequencing targeted [https://synapse.patsnap.com/article/how-does-whole-genome-sequencing-differ-from-targeted-sequencing]
- 11. Read length versus Depth of Coverage for Viral... | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0047046]
- 12. Targeted Sequencing Approach and Its Clinical Applications for the Molecular Diagnosis of Human Diseases - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9913990/]
- 13. A practical method to detect SNVs and indels from whole genome and exome sequencing data | Scientific Reports [https://www.nature.com/articles/srep02161]
- 14. Comparative evaluation of SNVs, indels, and structural variations detected with short- and long-read sequencing data | Human Genome Variation [https://www.nature.com/articles/s41439-024-00276-x]
- 15. Ultima Genomics UG 100 platform vs . Illumina NovaSeq X Series... [https://www.illumina.com/science/genomics-research/articles/ultima-ug-100-vs-illumina-novaseq-x-wgs-data.html]
- 16. Analysis of genetic differences underlying chilling stress tolerance... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-11986-z]
- 17. Determining Performance Metrics for Targeted Next Generation Sequencing Panels Using Reference Materials - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6172655/]
- 18. DNA Sequencing Costs: Data [https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data]
- 19. Next-Generation Sequencing Costs: The Sub $100 Genome [https://www.completegenomics.com/next-generation-sequencing-costs/]
- 20. The Cost of Sequencing a Human Genome [https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost]
- 21. The complete costs of genome sequencing: a microcosting study in cancer and rare diseases from a single center in the United Kingdom | Genetics in Medicine [https://www.nature.com/articles/s41436-019-0618-7]
- 22. Summary of findings – cost-effectiveness of next generation sequencing - Next Generation DNA Sequencing: A Review of the Cost Effectiveness and Guidelines - NCBI Bookshelf [https://www.ncbi.nlm.nih.gov/books/NBK274079/]
- 23. of Cost a full human sequencing - Our World in Data genome [https://ourworldindata.org/grapher/cost-of-sequencing-a-full-human-genome]
- 24. Measuring Genome and its Health Impact Sequencing Costs [https://www.wipo.int/en/web/global-health/w/news/2025/measuring-genome-sequencing-costs-and-its-health-impact]
- 25. “The Cost of Sequencing a Human Genome will fall by over 50% by... [https://www.linkedin.com/pulse/cost-sequencing-human-genome-fall-over-50-2025-3billionpairs]
- 26. Cost of NGS | Comparisons and budget guidance [https://www.illumina.com/science/technology/next-generation-sequencing/beginners/ngs-cost.html]
- 27. next generation Targeted (NGS) | IDT sequencing [https://www.idtdna.com/pages/technology/next-generation-sequencing/dna-sequencing/targeted-sequencing]
- 28. of DNA Preparation for Illumina... Sequencing Libraries [https://www.thermofisher.com/vn/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/next-generation-sequencing/dna-sequencing-preparation-illumina.html]
- 29. Performance comparison of whole-genome sequencing platforms - PubMed [https://pubmed.ncbi.nlm.nih.gov/22178993/]
- 30. Sequencing platform and library preparation choices impact viral metagenomes | BMC Genomics | Full Text [https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-14-320]
- 31. Paired Comparison of Whole Genome Sequencing and Comprehensive Targeted Sequencing of Pancreatic Cancer Tissue - PubMed [https://pubmed.ncbi.nlm.nih.gov/39890186/?fc=None&ff=20250201205228&v=2.18.0.post9+e462414]
- 32. Pipeline: DNA-Seq Bioinformatics : Analysis ... - GDC Docs Whole [https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/DNA_Seq_Variant_Calling_Pipeline/]
- 33. Genomic reproducibility in the bioinformatics era | Genome Biology | Full Text [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03343-2]
- 34. Approaches for... | Thermo Fisher Scientific - HK Targeted Sequencing [https://www.thermofisher.com/hk/en/home/life-science/sequencing/sequencing-learning-center/next-generation-sequencing-information/ngs-basics/targeted-sequencing-approaches.html]
- 35. . Amplicon : The Sequencing ... vs Hybridization Capture Comparison [https://www.cd-genomics.com/diseasepanel/amplicon-sequencing-vs-hybridization-capture-the-comparison-of-target-enrichment-methods-for-disease-panel.html]
- 36. Target Enrichment [https://sequencing.roche.com/us/en/products/product-category/target-enrichment.html]
- 37. Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9318977/]
- 38. | IDT Hybridization capture vs amplicon sequencing [https://www.idtdna.com/page/support-and-education/decoded-plus/targeted-sequencing-hybridization-capture-or-amplicon-sequencing/]
- 39. Target Enrichment Solutions for NGS: Amplicon Sequencing and Hybrid Capture - Paragon Genomics [https://www.paragongenomics.com/target-enrichment/]
- 40. Evaluation of Hybridization Capture Versus Amplicon‐Based Methods for Whole‐Exome Sequencing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4832303/]
- 41. Making the Most of Your NGS Data: Understanding Metrics for Target-enriched NGS [https://sequencing.roche.com/us/en/article-listing/metrics-for-target-enriched-ngs.html]
- 42. Comparison of Commercially Available Target Enrichment Methods for Next-Generation Sequencing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3605921/]
- 43. Targeted DNA-seq and RNA-seq of Reference Samples with Short-read and Long-read Sequencing | Scientific Data [https://www.nature.com/articles/s41597-024-03741-y]
- 44. Record-Breaking DNA Sequencing Technology Could Transform Newborn Care | The Scientist [https://www.the-scientist.com/record-breaking-dna-sequencing-technology-could-transform-newborn-care-73696]
- 45. World Record Set For Fastest Genome Sequencing [https://www.usnews.com/news/health-news/articles/2025-10-17/world-record-set-for-fastest-genome-sequencing]
- 46. Types of NIPT Technology | Whole - genome sequencing vs targeted [https://emea.illumina.com/clinical/reproductive-genetic-health/nipt/labs/nipt-technology-types.html]
- 47. and Exome Whole Market Genome ... Sequencing Research 2025 [https://finance.yahoo.com/news/whole-genome-exome-sequencing-market-090100270.html]
- 48. Targeted Sequencing And Resequencing Market Size, Share & 2030 Growth Trends Report [https://www.mordorintelligence.com/industry-reports/targeted-sequencing-and-resequencing-market]
- 49. Shaping the Future of Precision Oncology: A Comparative Analysis of... [https://www.meridianbioscience.com/lifescience/webinars-precision-oncology-may-2025/]
- 50. Whole genome sequencing - PubMed [https://pubmed.ncbi.nlm.nih.gov/20238084/]
- 51. xGen Amplicon Panels for NGS Oncology research | IDT [https://www.idtdna.com/pages/products/next-generation-sequencing/workflow/xgen-ngs-amplicon-sequencing/predesigned-amplicon-panels/oncology-inherited-disease-amp-panel]
- 52. Advances in Whole Genome Sequencing: Methods, Tools, and Applications in Population Genomics [https://www.mdpi.com/1422-0067/26/1/372]
- 53. NGS Panels for Hematological Cancer Profiling | Paragon Genomics [https://www.paragongenomics.com/applications/oncology/hematological-cancer-profiling/]
- 54. Frontiers | Bioinformatic challenges for pharmacogenomic study: tools for genomic data analysis [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1548991/full]
- 55. Recent advances in the methods and clinical applications of next-generation sequencing in genomic profiling and precision cancer therapy - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11830917/]
- 56. 2025 Trends: NGS [https://www.genengnews.com/topics/genome-editing/2025-trends-ngs/]
- 57. Next Generation Sequencing Market Surges 21.74% CAGR by 2034 [https://www.towardshealthcare.com/insights/next-generation-sequencing-market-sizing]
- 58. Genomic Biomarkers Market Surges Toward USD 17B by 2033 [https://media.market.us/genomic-biomarkers-market-news-2025/]
- 59. Gene Targeted Panels Sequencing [https://emea.illumina.com/techniques/sequencing/dna-sequencing/targeted-resequencing/targeted-panels.html]
- 60. Frontiers | Genomics costing tool: considerations for improving cost-efficiencies through cross scenario comparison [https://www.frontiersin.org/journals/public-health/articles/10.3389/fpubh.2024.1498094/full]
- 61. Adaptive sampling: redefining targeted sequencing [https://nanoporetech.com/blog/adaptive-sampling-redefining-targeted-sequencing]
- 62. DNA Sequencing: How to Choose the Right Technology - Front Line Genomics [https://frontlinegenomics.com/dna-sequencing-how-to-choose-the-right-technology/]
- 63. Performance evaluation of structural variation detection using DNBSEQ whole-genome sequencing | BMC Genomics | Full Text [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-11494-0]
- 64. Optimized high-throughput whole-genome sequencing workflow for surveillance of influenza A virus | Genome Medicine | Full Text [https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-025-01512-x]
- 65. Whole-Genome Sequencing (WGS) [https://www.illumina.com/techniques/sequencing/dna-sequencing/whole-genome-sequencing.html]
- 66. coverage and breadth of coverage Sequencing [https://www.reneshbedre.com/blog/sequencing-coverage.html]
- 67. DNA Sequencing Methods | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/sequencing/sequencing-learning-center/next-generation-sequencing-information/ngs-basics/dna-sequencing-methods.html]
- 68. Comparison of Commercially Available Target Enrichment Methods for Next-Generation Sequencing - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3605921/]
- 69. Rapid and highly-specific generation of targeted DNA sequencing libraries enabled by linking capture probes with universal primers | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0208283]
- 70. Top Whole (WGS) Companies & How to... Genome Sequencing [https://www.linkedin.com/pulse/top-whole-genome-sequencing-wgs-companies-how-qtxjc]
- 71. Performance comparison of whole-genome sequencing platforms - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4076012/]
- 72. Comparison of targeted next-generation sequencing for whole-genome sequencing of Hantaan orthohantavirus in Apodemus agrarius lung tissues | Scientific Reports [https://www.nature.com/articles/s41598-019-53043-2]
- 73. Market Size, Share & Forecast to 2032 Targeted Sequencing [https://www.researchandmarkets.com/report/targeted-sequencing]
- 74. Prioritizing non-coding regions based on human genomic constraint and sequence context with deep learning | Nature Communications [https://www.nature.com/articles/s41467-021-21790-4]
- 75. Complete Genomics Highlights Clinical Research Sequencing Solutions and Partnerships at AMP 2025 [https://www.prnewswire.com/news-releases/complete-genomics-highlights-clinical-research-sequencing-solutions-and-partnerships-at-amp-2025-302610501.html]
- 76. regionalpcs improve discovery of DNA methylation associations with complex traits | Nature Communications [https://www.nature.com/articles/s41467-024-55698-6]
- 77. 2025 and Beyond: The Future of Genomic Data Analysis and Innovations in Genomics Services [https://blog.crownbio.com/2025-and-beyond-the-future-of-genomic-data-analysis-and-innovations-in-genomics-services]
- 78. Best practices for the interpretation and reporting of clinical whole genome sequencing | npj Genomic Medicine [https://www.nature.com/articles/s41525-022-00295-z]
- 79. Target-Enhanced Whole-Genome Sequencing Shows Clinical Validity Equivalent to Commercially Available Targeted Oncology Panel - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12016826/]
- 80. Comparative assessment of the Sikun 2000 sequencing platform for... [https://www.nature.com/articles/s41598-025-04170-6?error=cookies_not_supported]
- 81. Evaluation of whole exome sequencing by targeted gene sequencing and Sanger sequencing - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0009898117302413]
- 82. Clinical sequencing: is WGS the better WES? - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4757617/]
- 83. Targeted next generation sequencing as a tool for precision medicine | BMC Medical Genomics | Full Text [https://bmcmedgenomics.biomedcentral.com/articles/10.1186/s12920-019-0527-2]
- 84. The performance of whole bisulfite genome on... sequencing [https://pmc.ncbi.nlm.nih.gov/articles/PMC10245168/]
- 85. of Comparison with Clinical Whole ... Genome Sequencing Targeted [https://www.researchsquare.com/article/rs-6993200/v1]
- 86. Bioinformatics Workflow for Whole Genome Sequencing - CD Genomics [https://www.cd-genomics.com/bioinformatics-workflow-for-whole-genome-sequencing.html]
- 87. analysis for Whole and... genome sequencing cancer genomics [https://pmc.ncbi.nlm.nih.gov/articles/PMC5834793/]
- 88. Rare Disease Targeted Sequencing [https://www.tst-web.illumina.com/areas-of-interest/genetic-disease/rare-disease-genomics/targeted-rare-disease-seq.html]
- 89. Opportunities and challenges of whole-genome and -exome sequencing | BMC Genomic Data | Full Text [https://bmcgenomdata.biomedcentral.com/articles/10.1186/s12863-017-0479-5]
- 90. Targeted Sequencing | Focus on key genes and regions of interest [https://www.illumina.com/techniques/sequencing/dna-sequencing/targeted-resequencing.html]
- 91. Whole genome and exome sequencing reference datasets from a multi-center and cross-platform benchmark study | Scientific Data [https://www.nature.com/articles/s41597-021-01077-5]